Abstract
Background
Flathead fishes of the genus Platycephalus are economically important demersal fishes that widely inhabit the continental shelves of tropical and temperate sea waters. This genus has a long history of taxonomic revision, and recently four Platycephalus species (Platycephalus sp. 1, Platycephalus sp. 2, P. indicus, and P. cultellatus) in the northwestern Pacific Ocean (NWP) have been recognized and redescribed. However, many aspects of their systematics and evolutionary history are unclear.
Results
A total of 411 individuals were sampled from 22 different sites across their distributions in the NWP. Three mitochondrial loci were sequenced to clarify the phylogeny and phylogeographic history of the fishes. The results showed significant differentiation of four Platycephalus species in the NWP with well-supported clades in which Platycephalus sp. 1 and Platycephalus sp. 2 were the closest, clustered with P. cultellatus, while their genetic relationship with P. indicus was the furthest. There were significant genealogical branches corresponding to P. indicus but not to other Platycephalus lineages. We further examined the phylogeographic patterns of 16 Platycephalus sp. 1 populations along the coastlines of China and Japan. A total of 69 haplotypes were obtained, with 23 shared among populations. One dominant haplotypic group, with a modest lineage structure and low levels of haplotype diversity and nucleotide diversity, was observed among Platycephalus sp. 1 populations. The demographic history reconstruction suggested a Platycephalus sp. 1 population expansion event dating back to the late Pleistocene.
Conclusions
Distributional rang variations may be the crucial factors sha** the genetic relationships of the genus Platycephalus. Reproductive schooling and potential egg/larval dispersal ability, coupled with the effects of ocean currents, are responsible for the present phylogeographic pattern of Platycephalus sp. 1.
Similar content being viewed by others
Background
In marine environments, the phylogeography of organisms may be shaped by complex interactions of extrinsic ecological factors (climatic changes, water temperature, ocean currents, anthropogenic influences, etc.) and intrinsic life-history characteristics (dispersal capacity, pelagic larval duration, etc.) [1, 2]. Moreover, the phylogeography of species can be demonstrated by study of species genetic diversity, population genetic structures and demographic history [1, 3].
During glacial cycles, continental shelves became alternately exposed and submerged under sea water, driving demographic extinction or expansion in numerous marine species [4]. In the Pleistocene glacial period, environmental changes were magnified in the marginal seas of the western Pacific Ocean [5]. With its distinct geographical features of various marginal seas, wide latitudinal range, and complex geological history, the northwestern Pacific (NWP) is an excellent natural region for investigating the genetic consequences of Pleistocene glaciation on population structures and geographical differentiation of marine fish species [6]. The Chinese coastal seas, including the Bohai Sea, Yellow Sea, East China Sea, and South China Sea, are relatively young marginal seas of the western Pacific Ocean. The life-history characteristics of marine fish species, their complex habitats, and seasonally changing current systems in Chinese coastal seas [7], could interact to promote, maintain, or homogenize genetic divergence after glacial periods [8, 9]. Accordingly, marine organisms inhabiting Chinese coastal seas are vital materials for studying the relationship between population genetic structures and geographical distributions [10,11,12].
Flathead fishes of genus Platycephalus, belonging to Family Platycephalidae, Order Scorpaeniformes, are widely distributed in tropical and temperate areas of the Indo-Western Pacific (IWP) and eastern Mediterranean [13,14,15,16,17]. They mainly inhabit estuarine and coastal seas to the edge of the continental shelf. With eyes on the upper surface of flattened heads, they often rest on the sea bottom and engage in ambush hunting. Flatheads have long been economically important fishes in some parts of east Asia and Australia [16]. They were among the top fishes to be commercially targeted by the Australian trawling industry [18]. The taxonomy of flatheads has a long history of confusion; approximately 150 species names have been proposed, but only 77 are considered valid [15,16,17, 19]. Cryptic species of flatheads have been recognized only recently, mostly in large expanses of the tropical IWP, such as in Australian waters [14, 17, 19, 20]. For example, a phylogenetic analysis of the genus Platycephalus with COI barcoding by Puckridge et al. [17] demonstrated a high level of genetic diversity of P. indicus with eight separated lineages distributed across large expanses of the IWP. The possibility of further cryptic speciation in this and other apparently widely distributed flatheads in the IWP needs to be considered.
There has long been only a single species of Platycephalus, P. indicus, recorded and studied in the NWP [21, 22]. Recently, newly recognized Platycephalus sp. 1 in China and Platycephalus sp. 2 in Japan were documented based on morphological characters and DNA barcoding, proving the validity of these species at the genetic level [21,22,23,24]. This recognition may coincide with the cryptic speciation of this widely distributed flathead species in the IWP. Meanwhile, P. cultellatus and P. indicus were also redescribed from the northern South China Sea [22, 25]. All four Platycephalus species distributed in the NWP are very similar in morphological and meristic values, with almost all characters overlap** [21, 22, 25]. This similarity leads to frequent misidentification of Platycephalus species in the NWP by regional ichthyologists. Given the glacial cycles mentioned above and the recent anthropogenic pressures, the phylogeographic history of the four Platycephalus species recorded in the NWP remains unclear. Moreover, no systematic research has been performed to study the population genetics of the newly recognized Platycephalus sp. 1, which is the only species widely distributed along Chinese coastal seas.
In the present study, populations of the genus Platycephalus were sampled along the coasts of China and Japan, as well as from extra-IWP reference locations. The specimens were identified as four species to investigate the phylogeny and evolutionary history of the genus Platycephalus. The phylogenetic relationships were reconstructed with multiple mitochondrial loci. These data were also used to characterize the population structure of the Platycephalus sp. 1 throughout its range. The population history of Platycephalus sp. 1 was inferred, and the results were used to illuminate how this species responded to the severe climatic fluctuations in the Pleistocene ice ages. Such information may assist in fisheries and ecosystem conservation and investigations of cryptic speciation in the genus Platycephalus in the NWP.
Results
Sequence variation within the genus Platycephalus
We obtained 651-bp COI sequences and 402-bp Cyt b sequences from 151 specimens of 22 populations of the four Platycephalus species (Fig. 1 and Additional file 1: Table S1). Comparison of the concatenated COI and Cyt b sequences revealed 57 distinct haplotypes among the four species, with a haplotype diversity (h) of 0.91 ± 0.02. These haplotypes were not shared among species. The numbers of haplotypes observed within each species were 30 for Platycephalus sp. 1, 16 for P. indicus, six for P. cultellatus, and five for Platycephalus sp. 2. Moreover, P. indicus exhibited the highest level of haplotype diversity (h = 0.90 ± 0.03), and P. cultellatus exhibited the lowest haplotype diversity (h = 0.60 ± 0.15), with Platycephalus sp. 1 (h = 0.76 ± 0.05) and Platycephalus sp. 2 (h = 0.71 ± 0.14) in between. The most common haplotype in Platycephalus sp. 1 (P1SH1) was observed in 46.6% of the 16 sampled Platycephalus sp. 1 populations, and most of the other haplotypes (72.4%) exhibited only one nucleotide difference.
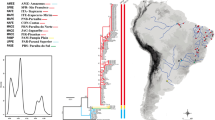
Sample locations for fishes of the genus Platycephalus and contemporary marine currents of the NWP. Samples are marked by abbreviations that correspond to those in Table 1 and Additional file 1: Table S1. KS: Kuroshio Current; TWC: Taiwan Warm Current; TSWC: Tsushima Warm Current; YSWC: Yellow Sea Warm Current; SBCC: Subei Coastal Current; MZCC: Minzhe Coastal Current; GDCC: Guangdong Coastal Current. The coastline map was originally made with Natural Earth (http://www.naturalearthdata.com/) and modified with marine currents according to Xu et al. [6]
Phylogenetic relationships of the genus Platycephalus
The haplotypes from concatenated COI and Cyt b sequences were used to infer the phylogenetic relationships of the sampled Platycephalus species. The concatenated tree topologies from the Bayesian and ML phylogenies were generally congruent (Fig. 2a and Additional file 2: Figure S1). The Bayesian tree (Fig. 2a) revealed a well-resolved phylogeny that included clades for four species. Generally, Platycephalus sp. 1 was closest to Platycephalus sp. 2, and the two species clustered with P. cultellatus, finally the three species clustered with P. indicus (Fig. 2a). The topology of the tree was shallow, with no significant genealogical structure regarding sampling locations for Platycephalus sp. 1, Platycephalus sp. 2, or P. cultellatus, respectively, whereas there was an obvious genealogical structure associated with sampling locations among populations of P. indicus. A significant lineage structure was distinguished among P. indicus of Okinawa, Beihai, and the Persian Gulf (Bahrain and Kuwait).
COI sequences from other Platycephalus species were utilized to assess the phylogeny and taxonomy of the genus Platycephalus; 510-bp COI sequences were collected from GenBank and this study (Additional file 3: Table S2). Generally, the phylogenetic relationships among the four NWP Platycephalus species considered above were identical to that of the concatenated tree, while there were additional Platycephalus species from Australia that were closely related to each other (Fig. 2b and Additional file 4: Figure S2). There were 67 sequences originally named “P. indicus” from GenBank; however, haplotype and phylogenetic analyses indicated that 27 of the 67 sequences actually belonged to Platycephalus sp. 1 (haplotypes P1SH1, P1SH4, and PIRC), covering samples from Korea, Rongcheng (China), Taiwan (China), and India. Moreover, 12 of the 67 sequences actually belonged to P. cultellatus (haplotypes PCSH1 and PCBH3) from the South China Sea, and one sequence belonged to Platycephalus sp. 2 from Japan (haplotype P2SH1) (Additional file 3: Table S2).
Lineage structures could be distinguished among geographic locations for populations of P. indicus. For example, the P. indicus populations from Taiwan (haplotype PITW) and east of Java (haplotype PV5) were more closely related to the population from Okinawa, and they further clustered with populations from the northern South China Sea, while the P. indicus samples from Australia, the Persian Gulf, and the Mediterranean were grouped as their own distinct clusters (Fig. 2b).
Population structure of P. indicus
The genetic relationships among the four P. indicus populations (n = 38) was quantified as corrected average pairwise differences, ΦST values, and AMOVA test results. Except for that between the BA and KU populations from the Persian Gulf, other pairwise values all exhibited large and significant divergence among populations from the South China Sea, Okinawa, and the Persian Gulf, with ΦST values ranging from 0.7794 to 0.9495 (Table 2A). An AMOVA among three geographically isolated groups of the South China Sea, Okinawa, and the Persian Gulf revealed a high but nonsignificant ΦCT value (ΦCT = 0.93, P = 0.1593), with 93.3% of the total genetic variation partitioned among groups. There was a high and significant ΦST (ΦST = 0.93, P = 0.000) for P. indicus populations. Scatter plotting from a Mantel test indicated a significant relationship (P = 0.0398) between FST/(1-FST) and geographic distance among the four P. indicus sample sites (Fig. 3a), indicating isolation by distance, with geographic distance explaining 84.9% of the variation in genetic differentiation for P. indicus (r = 0.92). The results indicated distance-related genetic differentiation among P. indicus populations. Considering the large geographic distance between populations as well as the limited sampling size for P. indicus, more samples across their intervening range are required to assess geographic patterns of genetic variation among P. indicus populations over their geographical expanse, i.e., whether gradual or with breaks at critical geographic sites.
Population structure of Platycephalus sp. 1
In order to gain more insights into the population and genealogical structure of the newly identified Platycephalus sp. 1 in the NWP, we amplified the mtDNA control region of additional specimens (n = 348) sampled from 15 Chinese coastal populations and one extra Tokyo Bay population from Japan (Table 1 and Fig. 1). There were 69 haplotypes detected among the 16 populations, and 46 haplotypes were unique and not shared among populations. Among the 23 haplotypes shared among populations, the most common haplotype (SH1) was observed in 156 (44.8%) individuals of the sampled populations, except for Tokyo Bay, Shantou and Zhuhai (Additional file 5: Table S3). Due to the limited number of samples and adjacent collection locations, the samples from Shantou, Shenzhen, Zhuhai, and Zhanjiang were combined and considered one “Guangdong” population in further population structure analyses (Table 1 and Fig. 1). The general haplotype diversity of Platycephalus sp. 1 populations was h = 0.78 ± 0.02, and the nucleotide diversity was π = 0.0041 ± 0.0026. The Nantong population exhibited the highest level of haplotype diversity (h = 0.97 ± 0.04), and the ** or similar morphological metrics, which resulted in frequent species misidentification, as shown by the few Platycephalus sequences in GenBank having been misidentified as P. indicus (Additional file 3: Table S2). Based on reports from our group and other studies, we summarized the recently described morphological characteristics of the four Platycephalus species along the Chinese coast and Japan, listed in Additional file 6: Table S4. All four species are characterized by 13 normal dorsal- and anal-fin rays, caudal fins with horizontal blackish bands, a scaled interorbital and occipital region, large caniniform teeth absent on the upper jaw, and a finger-like interopercular flap (Additional file 6: Table S4) [21,22,23, 25].
Among the four species upon which we focus, P. indicus is the only species with a single, anterior, small isolated spine on the first dorsal fin and a yellow marking in the middle of the caudal fin, while the other three species normally have two small isolated spines interior to the first dorsal fin and no yellow color on the caudal fin [21,22,23, 25]. The number of gill rakers and pored lateral line scales can also distinguish the species (Additional file 6: Table S4). The morphological information coupled with DNA barcoding can effectively aid in explicit species taxonomy and avoid misidentification and erroneous distributional records within Platycephalus in the NWP.
Phylogeny of the genus Platycephalus in the NWP
The phylogenetic relationships revealed distinct genetic structural differences among the sampled Platycephalus species from the NWP (Fig. 2). Platycephalus sp. 1 and Platycephalus sp. 2 were the most closely related, and these two species clustered with P. cultellatus, while the genetic relationship between P. indicus and the former three species was the furthest. The distributions of sampled Platycephalus species along the Chinese coast were sympatric or parapatric. Platycephalus sp. 1 was widely distributed along the Chinese coast, ranging from the Bohai Sea (Dalian) to the South China Sea (Beihai), whereas Platycephalus sp. 2 was geographically restricted to the coastal seas of Japan. This finding was consistent with those of reports from Japanese ichthyologists [23, 24, 27] and may be due to their specific thermal adaptation or geographic isolation by large expanses and ocean currents. Meanwhile, the distributions of P. indicus and P. cultellatus in China were confined to only the northern South China Sea. The abundant distribution of Platycephalus sp. 1 and the infrequent presence of P. indicus and P. cultellatus along the Chinese coast sharply contrasted, which suggested that the latter two species might prefer warmer water. The phylogeny was very shallow, with no clear phylogeographic patterns for Platycephalus sp. 1, Platycephalus sp. 2, and P. cultellatus populations in the NWP. However, a significant lineage structure related to geographical distribution was observed for P. indicus populations of Okinawa, Beihai, and the Persian Gulf (Fig. 2 and 3a). This structuring was probably because of obstacles that isolated the respective P. indicus populations, such as a large geographical expanse between the Persian Gulf and the other two populations (Fig. 3a), and the strong Kuroshio Current which isolates the Okinawa and Beihai populations (Fig. 1). These geographic and current barriers may have driven P. indicus populations to become taxonomically distinct as subspecies or even species. This inference coincided with the results of a previous study [17] in which eight divergent P. indicus lineages were designated, including P. indicus (China/Korea), P. indicus (Japan), P. cf. indicus (Hongkong), and P. indicus (Persian Gulf), which may indicate cryptic speciation. Regarding results of the Puckridge et al. study [17], the newly described Platycephalus sp. 1 and Platycephalus sp. 2 may correspond to the P. indicus (China/Korea) and P. indicus (Japan) lineages, respectively. Analysis of more samples across the intervening range is warranted to assess whether there might be gradual variation in P. indicus over a geographical expanse.
Geographic boundaries formed during the Pleistocene ice age have also played an important role in species dispersion. The genus Platycephalus is widely distributed in tropical and temperate areas of the IWP. This distribution suggests that the lineage could have originated in the tropical region, then dispersed north and became isolated in the NWP during glacial cycles. During the last glacial maximum (LGM), various refugia emerged, and geographic marine obstacles contributed to genetic isolation, which drove the differentiation that resulted in lineage structure divergence related to geographic distribution. For example, P. cultellatus was sampled only in the northern South China Sea, whose glacial refuge was the central South China Sea during the LGM. After the LGM, seasonal changes in the coastal currents in the northern South China Sea and the long developmental period of pelagic larvae contributed to the dispersion of the species.
In summary, distributional range variations, probably resulting from the divergence of thermal adaptation and the natural history effects of glaciation, may be the crucial drivers that shaped the genetic relationships of Platycephalus species in the NWP. Intrinsic life-history divergence may also have contributed to speciation processes, and hence further investigation into the life-history characteristics of these species is warranted.
Pleistocene glaciation and patterns of demographic history
Climatic condition during the Pleistocene ice age had dramatic effects on species distributions and abundance in the NWP [10, 28, 29]. Eurythermic species may have occupied various refugia during the glacial periods, which contributed to within-species divergence, while noneurythermic species may have been confined in limited refugia resulting in genetic homogeneity. However, the diverse life histories of marine species and the fluctuating environments (currents, sea level, water temperature, etc.) may have changed the phylogeographical scenario of fishes after their expansion from various Pleistocene refugia. The sequence mismatch distribution pattern of Platycephalus sp. 1 populations (Fig. 5) was unimodal and fully consistent with a population expansion scenario. Platycephalus sp. 1 populations also displayed a genetic pattern typical of a recent population expansion event, showing a single common haplotype (SH1) across most of its distribution, as well as a shallow star-shaped haplotype network (Fig. 4). Platycephalus sp. 1 was widespread along the China coastal seas, but during the LGM, the sea level fell approximately 120 to 140 m [30]. The sea level decline resulted in the exposure of the Yellow Sea and the Bohai Sea, the East China Sea was drawn back to the Okinawa Trough [31], and the South China Sea was separated from the Indian Ocean as a semi-closed sea [32]. Consequently, under the challenging environmental conditions in the glacial periods, Platycephalus sp. 1 may have experienced a reduction in population size over large parts of its distribution and survived in various isolated glacial refugia. The geographic isolation of these refugia contributed to genetic divergence among populations, quite possibly with adaptation to local conditions. During the interglacial stage, sea level and temperature rose, which may have contributed to the subsequent dispersal and demographic expansion of Platycephalus sp. 1 into the surrounding seas, such as the Yellow Sea, East China Sea, and South China Sea [33,34,35]. This interpretation also has been suggested for other marine species from this area [36, 37]. However, the data of the present study indicated only a modest population genetic structure, with one dominant haplotype and low levels of haplotype diversity and nucleotide diversity in Platycephalus sp. 1. Noting their high fecundity and habit of reproductive schooling, fluctuating oceanographic conditions would have contributed to a tendency for “sweepstake reproduction” for Platycephalus sp. 1 populations. In addition, heavy fishery pressure on demographically small lineages also could contribute to genetic homogeneity, causing the SH1 haplotype to become the most frequent haplotype within Platycephalus sp. 1 populations. Screenings of nuclear markers are warranted for further investigation of this hypothesis.
Effects of ocean currents and pelagic larval duration on population structure
Marine fishes generally present low genetic differentiation across geographic regions because of their high dispersal potential with planktonic egg, larval, juvenile, and adult stages. This dispersal could be accompanied by the absence of physical barriers isolating the species among ocean basins or adjacent marginal seas [38,39,40]. In this study, genetic homogeneity was observed among most of the Platycephalus sp. 1 populations along the Chinese coasts, while gene flow was limited only between Tokyo Bay samples and those from China (most of the ΦST > 0.3, P < 0.05) (Table 2B). Platycephalus has pelagic larvae that can live in the coastal seas before settling to the seabed as juveniles [41]. The larval duration period is not clear for most Platycephalus species. However, larvae of Platycephalus fuscus have been found in sea water up to two months after their breeding season [42]. Accordingly, high egg, larvae, or juvenile dispersal potential could be present in Platycephalus sp. 1 as well, and this might be an important factor promoting gene flow and homogeneity of populations across large geographic distances.
Marine current dynamics also affect species dispersal. The NWP has complex oceanic current systems along its continental margins, and seasonal current circulations are mostly driven by monsoon winds, for example, leading to water exchanges between the East China Sea and South China Sea through the Taiwan Strait (Fig. 1). Planktonic Platycephalus eggs or larvae could be transported northward during summer from the South China Sea to the East China Sea and the Yellow Sea by the Taiwan Warm Current and the Yellow Sea Warm Current [43]. On the other hand, with the winter monsoon winds, coastal waters streaming southward through the Taiwan Strait could drive extensive gene flow among populations along the China coastal seas [44,45,46]. With these strong marine currents, eggs, larvae, or juveniles of Platycephalus sp. 1 can travel long distances despite their limited adult dispersal capabilities. Several other fish species from Chinese seas, such as Liza affinis and Sillago japonica, with pelagic early-life stages, also could be mixed by ocean currents and present genetic homogeneity among Chinese coastal populations [47, 48]. Moreover, the Tokyo Bay population, which is partially blocked from Chinese coastal populations, showed low but significant genetic divergence from Platycephalus sp. 1 populations of the Chinese coast. The Tokyo Bay population apparently receives limited recruitment from the Yellow Sea and the East China Sea, likely due to the isolation by the Japanese landmass as well as the strong Kuroshio Current and Tsushima Warm Current (Fig. 1). This coincides with the long-term climatological events that may have created a barrier between populations of the Japanese and Chinese seas.
Conclusion
The present study reconstructed the phylogeny and inferred the evolutionary history of the genus Platycephalus from the NWP. Mitochondrial DNA sequence analysis demonstrated significant differentiation among the four Platycephalus species sampled in the NWP. The genealogical structure was shallow for three Platycephalus species, but subclades were detected within P. indicus. There was low genetic diversity and homogeneity in Platycephalus sp. 1 populations in the Chinese coastal seas. The demographic history of Platycephalus sp. 1 suggested a population expansion event dating to the late Pleistocene. Reproductive schooling and high larval dispersal coupled with the complex ocean currents are likely responsible for the present phylogeographic pattern of Platycephalus sp. 1. When managing Platycephalus fisheries, information that we developed can contribute to maintaining sustainable fisheries, improving ecosystem conservation, and investigating cryptic organismal evolution.
Methods
Sample collection
Specimens of the genus Platycephalus were collected from 22 locations (Table 1, Additional file 1: Table S1, and Fig. 1): Platycephalus sp. 1 were sampled from 15 sites along the Chinese coast and one site from Tokyo Bay, Japan. Platycephalus sp. 2 were collected from three sites in Japan. P. cultellatus were sampled from four sites in the northern South China Sea, and P. indicus were collected from Okinawa, Beihai, and the Persian Gulf (Bahrain and Kuwait). The samples were collected by trawl net in offshore waters from local fishermen or from fish market of local fishing ground, thus ensuring that the samples collected were representative of the local populations. Detailed information about the locations and numbers of specimens collected is listed in Additional file 1: Table S1. All individuals were identified on the basis of morphological characteristics [21, 22, 25], and a piece of muscle was taken from each individual and preserved in 95% ethanol for DNA extraction.
DNA extraction, PCR and sequencing analysis
Genomic DNA was extracted from muscle tissue by a standard proteinase K digestion and phenol-chloroform extraction method [49]. PCR amplifications were performed for three mitochondrial DNA (mtDNA) fragments: the control region (CR), cytochrome b (Cyt b), and cytochrome c oxidase I (COI). PCR was carried out in 25-μl reaction volumes with the primers listed in Additional file 7: Table S5. The PCR-amplified products were gel-separated, purified, sequenced with both primers, and analyzed on an ABI 3730 automated sequencer (Applied Biosystems, Waltham, MA, USA).
Sequence concatenations and file format conversions were implemented in Sequence Matrix [50]. Sequences were proofread, assembled, and aligned using DNASTAR software (DNASTAR, Inc., Madison, WI, USA). Haplotype identifications were determined in DnaSP [51]. Molecular diversity indices, such as the number of haplotypes, haplotype diversity (h), and nucleotide diversity (π), were obtained in ARLEQUIN [52].
Partition scheme and model selection
Various partition methods were employed to find the best-fit combination of substitution model parameters for the phylogenetic analysis. The best-fitting nucleotide substitution model was determined using the Bayesian approach implemented in jModelTest [53] for CR and in PartitionFinder 2 [54] for COI and Cyt b sequences. Protein-encoding sequences of COI and Cyt b were partitioned based on three codon positions, and the best codon substitution model was determined. To select the optimal model parameters and the best phylogenetic tree, all partition schemes were compared with the likelihood ratio test [55] for the maximum likelihood (ML) trees and Bayes factors (BFs) for the Bayesian trees. BFs were computed with a Step** Stone analysis [56, 57] in MrBayes [58]; the analysis included two MCMC runs with five chains and 100 steps of 1,000,000 generations each, sampling every 1,000 generations, and was stopped when it reached a mean standard deviation between split frequencies of less than 0.01.
Phylogeny reconstruction for the genus Platycephalus
To construct phylogenies based on the mtDNA sequences, six individuals were selected from each population of the sampled Platycephalus species. If the sampled specimens were fewer than six at a certain location, then all individuals were included. For phylogenetic analysis, 1) concatenated COI and Cyt b sequences of 151 specimens from populations of the four Platycephalus species were analyzed, and 2) homologous COI regions for other Platycephalus species were searched for using BLAST and downloaded from GenBank (https://www.ncbi.nlm.nih.gov/), excised and included in the combined COI phylogeny. The species and their COI-sequence GenBank accession numbers are listed in Additional file 3: Table S2. Based on the best-fit model estimated above, the ML tree was constructed with IQ-TERR [59], the Bayesian tree was constructed with MrBayes [58], and the neighbor-joining (NJ) tree was constructed with MEGA 6 [60] using the Kimura two-parameter (K2P) model [61] with 1,000 bootstrap replications. Five specimens of Cociella crocodile were used as an outgroup.
Population genetic structure analysis
Population structures based on the CR were analyzed for P. indicus and Platycephalus sp. 1, but not for P. cultellatus and Platycephalus sp. 1 due to their limited sample sizes. Pairwise population differences within P. indicus and Platycephalus sp. 1 populations were estimated with ΦST, and corrected average pairwise differences were calculated in ARLEQUIN. The statistical significance of these parameters was tested as table-wide significance (P < 0.05) with 10,000 permutations. Population structure was evaluated by a hierarchical analysis of molecular variance (AMOVA) [62] using 10,000 permutations in ARLEQUIN to examine the significant variation among groups of genetically similar populations. To evaluate hypothesized patterns of spatial genetic structures, three independent hierarchical AMOVAs were implemented across Platycephalus sp. 1 populations. One group was designated with all Platycephalus sp. 1 populations. Two groups (Pacific coast of Japan group - TO and Chinese coastal seas group) and four groups (Pacific coast of Japan group – TO; the Bohai and Yellow Sea group – DL, DY, WH, QD, NT; the East China Sea group - ZS, ND, CL, XM; and the South China Sea group – GD, BH, FC) were designated based on potential geographic isolation factors, such as the Tsushima Strait, Yangtze River estuary, and Taiwan Strait (Fig. 1). Moreover, independent hierarchical AMOVAs were implemented across P. indicus populations; three groups (the South China Sea group; the Okinawa group; the Persian Gulf group – KU, BA) were designated based on their geographic distance. Genetic split between populations was identified by finding the largest significant values of ΦCT among population groups. Haplotype networks were generally better than phylogenetic trees to describe population relationships [1]. The median joining haplotype network was estimated using Network [63], which was used for constructing large networks post-processed with parsimony [64].
To test for isolation by distance, pairwise values of FST/(1-FST) were plotted against geographical distance between sample sites of Platycephalus sp. 1 (one-dimensional step**-stone model), while pairwise values of FST/(1-FST) were plotted against the logarithm of distance between sample sites of P. indicus (two-dimensional step**-stone model) considering the wide distribution range of P. indicus. The strength and significance of the relationship between genetic distances and geographic distances was assessed using reduced major axis (RMA) regression and Mantel tests using the software ARLEQUIN.
Demographic history of Platycephalus sp. 1
The historical demographic pattern was examined for Platycephalus sp. 1 populations. The neutrality mutation test was calculated using Tajima’s D [65] and Fu’s FS [66], with significance tested using 1,000 replicates in ARLEQUIN. Historical demographic expansions also were tested with the mismatch distributions of all pairwise differences in ARLEQUIN for pure demographic [67] or spatial [68] expansions. Raggedness indices were estimated to evaluate the DNA sequence mismatch distributions of the respective populations [69], and the observed frequencies were compared to model predictions based on 10,000 permutations. If the observations were not significantly different from model predictions, the time of expansion was estimated by the equation Ƭ = 2 μt, where μ is the mutation rate [67].
BEAST v.1.7.4 was employed to estimate the changes in effective population size (Ne) across time and to create Bayesian skyline plots [70]. The unique haplotypes of total samples were run for 100,000,000 generations with a constant skyline model of 15 groups. The strict molecular clock model was the prior distribution, with an initial value of 0.06 and a range from 0.03 to 0.10 [10, 26]. To test convergence, trace plots were inspected in Tracer v1.6 [71], and the effective sample sizes (ESSs) were confirmed to exceed 200.
Availability of data and materials
All relevant data are within the paper and its additional files. We have submitted the mitochondrial DNA sequences to GenBank with accession numbers MH615133-MH615480, MH615481-MH615630, MH615631–615781.
Abbreviations
- COI :
-
Cytochrome c oxidase I
- CR :
-
Control region
- Cyt b :
-
Cytochrome b
- h :
-
Haplotype diversity
- IWP:
-
Indo-Western Pacific
- K2P:
-
Kimura two-parameter
- LGM:
-
Last glacial maximum
- ML:
-
Maximum likelihood
- mtDNA:
-
Mitochondrial DNA
- NWP:
-
Northwestern Pacific
- PCR:
-
Polymerase chain reaction
- π :
-
Nucleotide diversity
References
Klimova A, Phillips CD, Fietz K, Olsen MT, Harwood J, Amos W, Hoffman JI. Global population structure and demographic history of the grey seal. Mol Ecol. 2014;23:3999–4017.
Bayha KM, Chang MH, Mariani CL, Richardson JL, Edwards DL, Deboer TS, Moseley C, Aksoy E, Decker MB, Gaffney PM, Harbison GR, McDonald JH, Caccone A. Worldwide phylogeography of the invasive ctenophore Mnemiopsis leidyi (Ctenophora) based on nuclear and mitochondrial DNA data. Biol Invasions. 2015;17:827–50.
Da Silva R, Veneza I, Sampaio I, Araripe J, Schneider H, Gomes G. High levels of genetic connectivity among populations of yellowtail snapper, Ocyurus chrysurus (Lutjanidae-Perciformes), in the western South Atlantic revealed through multilocus analysis. PLoS One. 2015;10:e0122173.
Ludt WB, Rocha LA. Shifting seas: the impacts of Pleistocene Sea-level fluctuations on the evolution of tropical marine taxa. J Biogeogr. 2015;42:25–38.
Wang PX. Response of Western Pacific marginal seas to glacial cycles: paleoceanographic and sedimentological features. Mar Geol. 1999;156:5–39.
Xu SY, Song N, Zhao LL, Cai SS, Han ZQ, Gao TX. Genomic evidence for local adaptation in the ovoviviparous marine fish Sebastiscus marmoratus with a background of population homogeneity. Sci Rep. 2017;7:1562.
Liu JY. Status of marine biodiversity of the China seas. PLoS One. 2013;8:e50719.
Jue NK, Brule T, Coleman FC, Koenig CC. From shelf to shelf: assessing historical and contemporary genetic differentiation and connectivity across the Gulf of Mexico in gag, Mycteroperca microlepis. PLoS ONE. 2015;10:e0120676.
Wang J, Tsang LM, Dong YW. Causations of phylogeographic barrier of some rocky shore species along the Chinese coastline. BMC Evol Biol. 2015;15:114.
Liu JX, Gao TX, Wu SF, Zhang YP. Pleistocene isolation in the northwestern Pacific marginal seas and limited dispersal in a marine fish, Chelon haematocheilus (Temminck & Schlegel, 1845). Mol Ecol. 2007;16:275–88.
He LJ, Zhang AB, Weese D, Li SF, Li JS, Zhang J. Demographic response of cutlassfish (Trichiurus japonicus and T. nanhaiensis) to fluctuating palaeo-climate and regional oceanographic conditions in the China seas. Sci Rep. 2014;4:6380.
Zhang YH, Pham NK, Zhang HX, Lin JD, Lin Q. Genetic variations in two seahorse species (Hippocampus mohnikei and Hippocampus trimaculatus): evidence for middle Pleistocene population expansion. PLoS One. 2014;9:e105494.
Hureau JC. Platycephalidae. In: Whitehead PJP, Bauchot ML, Hureau JC, Nielsen J, Tortonese E, et al.. Fishes of the North-eastern Atlantic and the Mediterranean, Volume III. UNESCO, Paris. 1986; 1241–1242.
Shao KT, Chen JP. Fishes of the family Platycephalidae (Teleostei: Platycephaloidei) of Taiwan with descriptions of two new species. Bull Inst Zool Acad Sinica. 1987;26:77–94.
Imamura H. Phylogeny of the family Platycephalidae and related taxa (Pisces: Scorpaeniformes). Species Diversity. 1996;1:123–233.
Knapp LW. Platycephalidae. In: Carpenter KE, Niem VH, editors. FAO species identification guide for fishery purposes. The Living Marine Resources of the Western Central Pacific, Volume 4, Bony Fishes, part 2 (Mugilidae to Carangidae). Rome: FAO; 1999. p. 2385–421.
Puckridge M, Andreakis N, Appleyard SA, Ward RD. Cryptic diversity in flathead fishes (Scorpaeniformes: Platycephalidae) across the indo-West Pacific uncovered by DNA barcoding. Mol Ecol Resour. 2013;13:32–42.
Fairbridge WS. The over-fishing of the East Australian trawl fishery. Cronulla, N.S.W.: Indo-Pacific Fisheries Council Proceedings 2nd Meeting; 1950.
Imamura H. Taxonomic revision of the flathead fish genus Platycephalus Bloch, 1785 (Teleostei: Platycephalidae) from Australia, with description of a new species. Zootaxa. 2015;3904:151–207.
Imamura H. Re-diagnosis of the marbled flathead, Platycephalus marmoratus (Actinopterygii: Teleostei: Platycephalidae), with comments on the composition of the type series. Species Divers. 2006;11:295–306.
Qin Y, Song N, Zou JW, Zhang ZH, Cheng GP, Gao TX, Zhang XM. A new record of a flathead fish (Teleostei: Platycephalidae) from China based on morphological characters and DNA barcoding. Chin J Oceanol Limnol. 2013;31(3):617–24.
Chen Z, Gao T. Morphological re-description and DNA barcoding of Platycephalus indicus collected from Beihai, China. Pakistan J Zool. 2017;49(5):1647–55.
Masuda Y, Haraguchi M, Ozawa T, Matsui S, Hayashi S. Morphological and biochemical comparisons of two flathead species of the genus Platycephalus collected from the Western Suonada and southern Yatsushiro seas, Japan. Nippon Suisan Gakkaishi. 1997;63:345–52.
Nakabo T. Fishes of Japan with Pictorial Keys to the Species, English edition. Tokyo: Tokai University Press; 2002.
Chen Z, Song N, Zou JW, Gao TX. Rediscovery and elaborate description of Platycephalus cultellatus Richardson, 1846 collected from the coastal waters of South China Sea. Aqua Living Res. 2018;31:32.
Li YF, Chen GB, Yu J, Wu SQ, **ong D, Li X, Cui K, Li YZ. Population genetics of Thamnaconus hypargyreus (Tetraodontiformes: Monacanthidae) in the South China Sea. Mitochondrial DNA. 2016;27:798–805.
Yamada U, Tokimura M, Horikawa H, Nakabo T. Fishes and Fisheries of the East China and Yellow Seas. Tokai University Press. (in Japanese) 2007.
Avise JC. Phylogeography: the history and formation of species. Cambridge, MA: Harvard University Press; 2000.
Dynesius M, Jansson R. Evolutionary consequences of changes in species’ geographical distributions driven by Milankovitch climate oscillations. Proc Natl Acad Sci U S A. 2000;97:9115–20.
Lambeck K, Esat TM, Potter EK. Links between climate and sea levels for the past three million years. Nature. 2002;419:199–206.
Wang PX. The ice-age China Sea status and problems. Quat Sci. 1990;2:111–24.
Voris HK. Maps of Pleistocene Sea levels in Southeast Asia: shorelines, river systems and time durations. J Biogeogr. 2000;27:1153–67.
Hearty PJ, Hollin JT, Neumann AC, Leary MJO, McCulloch M. Global Sea-level fluctuations during the last interglaciation (MIS 5e). Quat Sci Rev. 2007;26:2090–112.
Pillans B, Chappell J, Naish TR. A review of the Milankovitch climatic beat: template for Plio-Pleistocene Sea-level changes and sequence stratigraphy. Sediment Geol. 1998;122:5–21.
Stirling CH, Esat TM, Lambeck K, McCulloch MT. Timing and duration of the last interglacial: evidence for a restricted interval of widespread coral reef growth. Earth Planet Sc Lett. 1998;160:745–62.
He LJ, Zhang AB, Weese D, Zhu CD, Jiang CJ, Qiao ZG. Late Pleistocene population expansion of Scylla paramamosain along the coast of China: a population dynamic response to the last interglacial sea level highstand. J Exp Mar Biol Ecol. 2010;385:20–8.
Ma CY, Cheng QQ, Zhang QY. Genetic diversity and demographical history of Coilia ectenes (Clupeiformes: Engraulidae) inferred from the complete control region sequences of mitochondrial DNA. Mitochondrial DNA. 2012;23:396–404.
Grant W, Bowen B. Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J Hered. 1998;89:415–26.
Hewitt GM. The genetic legacy of the quaternary ice ages. Nature. 2000;405:907–13.
Palumbi S. Genetic divergence, reproductive isolation, and marine speciation. Annu Rev Ecol Syst. 1994;25:547–72.
Syahailatua A, Roughan M, Suthers IM. Characteristic ichthyoplankton taxa in the separation zone of the east Australian current: larval assemblages as tracers of coastal mixing. Deep-Sea Research Part II: Topical Studies in Oceanography. 2011;58:678–90.
Gray CA, Miskiewicz AG. Larval fish assemblages in south-east Australian coastal waters: seasonal and spatial structure. Estuar Coast Shelf Sci. 2000;50:549–70.
Ichikawa H, Beardsley RC. The current system in the yellow and East China seas. J Oceanogr. 2002;58(1):77–92.
Hu J, Kawamura H, Hong H, Qi Y. A review on the currents in the South China Sea: seasonal circulation, South China Sea warm current and Kuroshio intrusion. J Oceanogr. 2000;56:607–24.
Guan BX, Fang GH. Winter counter-wind currents off the southeastern China coast: a review. J Oceanogr. 2006;62:1–24.
Tsang LM, Chan BKK, Ma KY, Chu KH. Genetic differentiation, hybridization and adaptive divergence in two subspecies of the acorn barnacle Tetraclita japonica in the northwestern Pacific. Mol Ecol. 2008;17:4151–63.
Ni G, Li Q, Kong LF, Yu H. Comparative phylogeography in marginal seas of the northwestern Pacific. Mol Ecol. 2014;23:534–48.
Liu L, Liu LQ, Gao TX, Song N. Phylogeographic pattern of Liza affinis populations in Chinese coastal waters: estimation of larval dispersal potential. Mitochondrial DNA Part A. 2018;29(8):1253–60.
Green MR and Sambrook J. Isolation of high-molecular-weight DNA from mammalian cells using proteinase K and phenol. Molecular Cloning: A Laboratory Manual, 4th ed. 2012. 47–53.
Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multigene datasets with character set and codon information. Cladistics. 2011;27(2):171–80.
Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2.
Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Resour. 2010;10:564–7.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772.
Lanfear R, Calcott B, Ho SYW, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–701.
Wilks S. The large sample distribution of the likelihood ratios for testing composite hypothesis. Ann Math Stat. 1938;9:60–2.
Fan Y, Wu R, Chen MH, Kuo L, Lewis PO. Choosing among partition models in Bayesian phylogenetics. Mol Biol Evol. 2011;28:523–32.
**e W, Lewis PO, Fan Y, Kuo L, Chen MH. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst Biol. 2011;60:150–60.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: effective Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42.
Nguyen LT, Schmidt HA, Von HA, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Kimura M. A simple method of estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–91.
Bandelt HJ, Forster P, Roh A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48.
Polzin T, Daneshmand SV. On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett. 2003;31:12–20.
Aris-Brosou S, Excoffier L. The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism. Mol Biol Evol. 1996;13:494–504.
Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–25.
Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–69.
Excoffier L. Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol. 2004;13:853–64.
Harpending HC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol. 1994;66:591–600.
Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214.
Rambaut A, Drummond AJ, **e D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018;67(5):901–4.
Acknowledgements
We sincerely appreciated the instruction of Dr. H. Imamura for Platycephalus identification. Thanks to L. Li and J. Zou for sample collection. Thanks for Y. Qin for specimen identification. Thanks for Dr. M. Hui for comprehensive comments.
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 41776171). The funding body had no role in the design of the study, collection, analysis and interpretation of data, or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
JC analyzed and interpreted the data, and wrote the manuscript; ZW performed the mtDNA laboratory experiments and analyzed the data; NS contributed to interpretation and discussion of the results; TY provided samples and interpreted the results; TG contributed to the manuscript’s design, discussion and interpretation of the results. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The field studies did not involve any endangered or protected species. Platycephalus is not protected by law. No fishing license was required for collection of samples from all locations. It is a commercially harvested species in China, and the fishes were already dead after being collected by trawling by local fishermen for commercial purposes. None of the authors was involved in the collection of the fishes. Animal Ethics Committee approval was not needed because no handing of live animals was involved.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Sample information and diversity metrics of the genus Platycephalus in the NWP. (DOCX 26 kb)
Additional file 2:
Figure S1. Phylogenetic relationships of the genus Platycephalus based on concatenated COI and Cyt b sequences. Partitioned maximum likelihood tree and codon maximum likelihood tree are on top and bottom, respectively. (TIF 1372 kb)
Additional file 3:
Table S2. COI sequences of the Platycephalus from GenBank. (DOCX 44 kb)
Additional file 4:
Figure S2. Phylogenetic relationships of the genus Platycephalus based on COI sequences from this study and from GenBank. Bayesian inference tree, partitioned maximum likelihood tree, and codon maximum likelihood tree are from left to right. (TIF 1953 kb)
Additional file 5:
Table S3. Distribution of haplotypes shared among populations of the Platycephalus sp. 1. (DOCX 17 kb)
Additional file 6:
Table S4. Comparison of selected morphological characters of four Platycephalus species from Chinese coastal seas and Pacific coast of Japan, as found in bibliography. (DOCX 20 kb)
Additional file 7:
Table S5. Primers information of mitochondrial DNA for the Platycephalus. (DOCX 19 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Cheng, J., Wang, Z., Song, N. et al. Phylogeographic analysis of the genus Platycephalus along the coastline of the northwestern Pacific inferred by mitochondrial DNA. BMC Evol Biol 19, 159 (2019). https://doi.org/10.1186/s12862-019-1477-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-019-1477-1