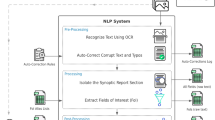
Abstract
Background
Longitudinal data on key cancer outcomes for clinical research, such as response to treatment and disease progression, are not captured in standard cancer registry reporting. Manual extraction of such outcomes from unstructured electronic health records is a slow, resource-intensive process. Natural language processing (NLP) methods can accelerate outcome annotation, but they require substantial labeled data. Transfer learning based on language modeling, particularly using the Transformer architecture, has achieved improvements in NLP performance. However, there has been no systematic evaluation of NLP model training strategies on the extraction of cancer outcomes from unstructured text.
Results
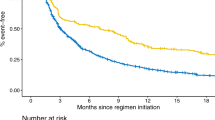
We evaluated the performance of nine NLP models at the two tasks of identifying cancer response and cancer progression within imaging reports at a single academic center among patients with non-small cell lung cancer. We trained the classification models under different conditions, including training sample size, classification architecture, and language model pre-training. The training involved a labeled dataset of 14,218 imaging reports for 1112 patients with lung cancer. A subset of models was based on a pre-trained language model, DFCI-ImagingBERT, created by further pre-training a BERT-based model using an unlabeled dataset of 662,579 reports from 27,483 patients with cancer from our center. A classifier based on our DFCI-ImagingBERT, trained on more than 200 patients, achieved the best results in most experiments; however, these results were marginally better than simpler “bag of words” or convolutional neural network models.
Conclusion
When develo** AI models to extract outcomes from imaging reports for clinical cancer research, if computational resources are plentiful but labeled training data are limited, large language models can be used for zero- or few-shot learning to achieve reasonable performance. When computational resources are more limited but labeled training data are readily available, even simple machine learning architectures can achieve good performance for such tasks.
Similar content being viewed by others
Background
Precision oncology, defined as tailoring cancer treatment to the individual clinical and molecular characteristics of patients and their tumors [1], is an increasingly important goal in cancer medicine. This strategy requires linking tumor molecular data [2] to data on patient outcomes to ask research questions about the association between tumor characteristics and treatment effectiveness. Despite the increasing sophistication of molecular and bioinformatic techniques for genomic data collection, the ascertainment of corresponding clinical outcomes from patients who undergo molecular testing has remained a critical barrier to precision cancer research. Outside of therapeutic clinical trials, key clinical outcomes necessary to address major open questions in precision oncology, such as which biomarkers predict cancer response (improvement) and progression (worsening), are generally recorded only in the free text documents generated by radiologists and oncologists as they provide routine clinical care.
Clinical cancer outcomes other than overall survival are not generally captured in standard cancer registry workflows; historically, abstraction of such outcomes from the electronic health record (EHR) has therefore required resource-intensive manual annotation. If this abstraction has occurred at all, it has generally been performed within individual research groups in the absence of data standards, yielding datasets of questionable generalizability. To address this gap, our research group developed the ‘PRISSMM’ framework for EHR review. PRISSMM is a structured rubric for manual annotation of each pathology, radiology/imaging, and medical oncologist report to ascertain cancer features and outcomes; each imaging report is reviewed in its own right to determine whether it describes cancer response, progression, or neither [3]. This annotation process also effectively yields document labels that can be used to train machine learning-based natural language processing (NLP) models to recapitulate these manual annotations. We previously detailed the PRISSMM annotation directives for ascertaining cancer outcomes and demonstrated the feasibility of using PRISSMM labels to train NLP models that can identify cancer outcomes within imaging reports [3, 4] and medical oncologist notes [4, 5].
While applying NLP to clinical documents can dramatically accelerate outcome ascertainment, training these models from randomly initialized weights remains resource-intensive, requiring thousands of manually annotated documents. Modern advances in NLP could reduce this data labeling burden. Semi-supervised learning techniques based on language modeling, or using components of a sentence or document to predict the remainder of the text, have become cornerstones of NLP [ The overall cohort for this analysis consisted of patients with cancer participating in a single-institution genomic profiling study [27], and relevant data consisted of imaging reports for each patient. Each report was treated as its own unit of analysis, and reports were divided, at the patient level, into training (80%), validation (10%), and test (10%) datasets. For language model pre-training on data from our institution, reports for all patients in the training set were included. This dataset included 662,579 reports from 27,483 patients with multiple types of cancer whose tumors were sequenced through our institutional precision medicine study [27]. For classification model training, the imaging reports for a subset of patients with lung cancer were manually annotated to ascertain the presence of cancer response or progression in each report using the PRISSMM framework, as previously described [3]. Briefly, during manual annotation, human reviewers recorded whether each imaging report indicated any cancer, and if so, whether it was responding/improving, progressing/worsening, stable (neither improving nor worsening), mixed (with some areas improving and some worsening), or indeterminate (if assigning a category was not possible due to radiologist uncertainty or other factors). For NLP model training, response/improvement and progression/worsening were each treated as binary outcomes, such that an imaging report indicating no cancer, or indicating stable, mixed, or indeterminate cancer status, was coded as neither improving nor worsening. This process, and interrater reliability statistics for manual annotation, have been described previously [3]. The classification dataset consisted of 14,218 labeled imaging reports for 1112 patients. Among the reports, 1635 (11.5%) indicated cancer response/improvement, and 3522 (24.8%) indicated cancer progression/worsening. Our baseline architecture was a simple logistic regression model in which the text of each imaging report was vectorized using term frequency-inverse document frequency (TF-IDF) vectorization [28]. This model used elastic net regularization with alpha = 0.0001, L1 ratio of 0.15, and was trained with stochastic gradient descent. Other architectures included one-dimensional convolutional neural networks (CNNs) [Methods
Cohort
Models
Availability of data and materials
The underlying EHR text reports used to train and evaluate NLP models for these analyses constitute protected health information for DFCI patients and therefore cannot be made publicly available. Researchers with DFCI appointments and Institutional Review Board (IRB) approval can access the data on request. For external researchers, access would require collaboration with the authors and eligibility for a DFCI appointment per DFCI policies. Scripts used to implement, train, and evaluate the models are deposited in the public repository https://github.com/marakeby/clinicalNLP2.
References
Garraway LA, Verweij J, Ballman KV. Precision oncology: an overview. J Clin Oncol Off J Am Soc Clin Oncol. 2013;31(15):1803–5.
AACR Project GENIE Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–31.
Kehl KL, Elmarakeby H, Nishino M, Van Allen EM, Lepisto EM, Hassett MJ, et al. Assessment of deep natural language processing in ascertaining oncologic outcomes from radiology reports. JAMA Oncol. 2019;5(10):1421–9.
Kehl KL, Xu W, Gusev A, Bakouny Z, Choueiri TK, Riaz IB, et al. Artificial intelligence-aided clinical annotation of a large multi-cancer genomic dataset. Nat Commun. 2021;12(1):7304.
Kehl KL, Xu W, Lepisto E, Elmarakeby H, Hassett MJ, Van Allen EM, et al. Natural language processing to ascertain cancer outcomes from medical oncologist notes. JCO Clin Cancer Inform. 2020;4:680–90.
Dai AM, Le QV. Semi-supervised sequence learning. ar**v; 2015 [cited 2022 Sep 6]. http://arxiv.org/abs/1511.01432
Howard J, Ruder S. Universal language model fine-tuning for text classification. ar**v; 2018 [cited 2022 Sep 6]. http://arxiv.org/abs/1801.06146
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN et al. Attention is all you need. ar**v; 2017 [cited 2022 Sep 6]. http://arxiv.org/abs/1706.03762
Devlin J, Chang MW, Lee K, Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding. ar**v; 2019 [cited 2022 Sep 6]. http://arxiv.org/abs/1810.04805
Huang K, Altosaar J, Ranganath R. ClinicalBERT: modeling clinical notes and predicting hospital readmission. ar**v; 2020 Nov [cited 2022 May 31]. Report No. http://arxiv.org/abs/1904.05342
Dai Z, Yang Z, Yang Y, Carbonell J, Le QV, Salakhutdinov R. Transformer-XL: Attentive language models beyond a fixed-length context. ar**v; 2019 [cited 2022 Sep 6]. http://arxiv.org/abs/1901.02860
Kitaev N, Kaiser Ł, Levskaya A. Reformer: the efficient transformer. ar**v; 2020 [cited 2022 Sep 6]. http://arxiv.org/abs/2001.04451
Beltagy I, Peters ME, Cohan A. Longformer: the long-document transformer. ar**v; 2020 [cited 2022 Sep 6]. http://arxiv.org/abs/2004.05150
Olthof AW, Shouche P, Fennema EM, IJpma FFA, Koolstra RHC, Stirler VMA, et al. Machine learning based natural language processing of radiology reports in orthopaedic trauma. Comput Methods Programs Biomed. 2021;208:106304.
Chaudhari GR, Liu T, Chen TL, Joseph GB, Vella M, Lee YJ, et al. Application of a domain-specific BERT for detection of speech recognition errors in radiology reports. Radiol Artif Intell. 2022;4(4): e210185.
Nakamura Y, Hanaoka S, Nomura Y, Nakao T, Miki S, Watadani T, et al. Automatic detection of actionable radiology reports using bidirectional encoder representations from transformers. BMC Med Inform Decis Mak. 2021;21(1):262.
Olthof AW, van Ooijen PMA, Cornelissen LJ. Deep learning-based natural language processing in radiology: the impact of report complexity, disease prevalence, dataset size, and algorithm type on model performance. J Med Syst. 2021;45(10):91.
Wei J, Bosma M, Zhao VY, Guu K, Yu AW, Lester B et al. Finetuned language models are zero-shot learners. ar**v; 2022 [cited 2023 May 26]. http://arxiv.org/abs/2109.01652
Raffel C, Shazeer N, Roberts A, Lee K, Narang S, Matena M, et al. Exploring the limits of transfer learning with a unified text-to-text transformer. ar**v; 2020 [cited 2023 May 22]. http://arxiv.org/abs/1910.10683
Chung HW, Hou L, Longpre S, Zoph B, Tay Y, Fedus W, et al. Scaling instruction-finetuned language models. ar**v; 2022 [cited 2023 May 22]. http://arxiv.org/abs/2210.11416
Gutiérrez BJ, McNeal N, Washington C, Chen Y, Li L, Sun H, et al. Thinking about GPT-3 in-context learning for biomedical IE? Think again. ar**v; 2022 [cited 2023 May 26]. http://arxiv.org/abs/2203.08410
Kim Y. Convolutional neural networks for sentence classification. ar**v; 2014 [cited 2022 Sep 6]. http://arxiv.org/abs/1408.5882
Cho K, van Merrienboer B, Gulcehre C, Bahdanau D, Bougares F, Schwenk H, et al. Learning phrase representations using RNN encoder–decoder for statistical machine translation. ar**v; 2014 [cited 2022 Sep 6]. http://arxiv.org/abs/1406.1078
Huang XS, Perez F, Ba J, Volkovs M. Improving transformer optimization through better initialization. In: Proceedings of the 37th international conference on machine learning. PMLR; 2020 [cited 2022 Sep 6]. p. 4475–83. https://proceedings.mlr.press/v119/huang20f.html
Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2019;btz682.
Lehman E, Jain S, Pichotta K, Goldberg Y, Wallace BC. Does BERT pretrained on clinical notes reveal sensitive data? ar**v; 2021 Apr [cited 2022 Jun 2]. Report No. http://arxiv.org/abs/2104.07762
Sholl LM, Do K, Shivdasani P, Cerami E, Dubuc AM, Kuo FC, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1(19): e87062.
Salton G, Buckley C. Term-weighting approaches in automatic text retrieval. Inf Process Manag. 1988;24(5):513–23.
Wolf T, Debut L, Sanh V, Chaumond J, Delangue C, Moi A, et al. HuggingFace’s transformers: state-of-the-art natural language processing. ar**v; 2020 [cited 2022 Sep 6]. http://arxiv.org/abs/1910.03771
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. PyTorch: an imperative style, high-performance deep learning library. ar**v; 2019 [cited 2022 Sep 6]. http://arxiv.org/abs/1912.01703
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. ar**v; 2016 [cited 2022 Sep 6]. http://arxiv.org/abs/1603.04467
Zhang S, Roller S, Goyal N, Artetxe M, Chen M, Chen S, et al. OPT: open pre-trained transformer language models. ar**v; 2022 [cited 2023 May 30]. http://arxiv.org/abs/2205.01068
Sanh V, Webson A, Raffel C, Bach SH, Sutawika L, Alyafeai Z, et al. Multitask prompted training enables zero-shot task generalization. ar**v; 2022 [cited 2023 May 30]. http://arxiv.org/abs/2110.08207
Lu Q, Dou D, Nguyen T. ClinicalT5: a generative language model for clinical text. In: Findings of the association for computational linguistics: EMNLP 2022. Abu Dhabi, United Arab Emirates: Association for Computational Linguistics; 2022 [cited 2023 May 30]. p. 5436–43. https://aclanthology.org/2022.findings-emnlp.398
Lehman E, Hernandez E, Mahajan D, Wulff J, Smith MJ, Ziegler Z, et al. Do we still need clinical language models? ar**v; 2023 [cited 2023 May 30]. http://arxiv.org/abs/2302.08091
Phan LN, Anibal JT, Tran H, Chanana S, Bahadroglu E, Peltekian A, et al. SciFive: a text-to-text transformer model for biomedical literature. ar**v; 2021 [cited 2023 May 30]. http://arxiv.org/abs/2106.03598
Loshchilov I, Hutter F. Decoupled weight decay regularization. 2017 Nov 14 [cited 2022 Sep 6]; https://arxiv.org/abs/1711.05101v3
Chicco D, Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics. 2020;21(1):6.
Johnson AEW, Pollard TJ, Shen L, Lehman L, Wei H, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3(1):160035.
Liu Y, Ott M, Goyal N, Du J, Joshi M, Chen D, et al. RoBERTa: a robustly optimized BERT pretraining approach. ar**v; 2019 [cited 2023 Jun 5]. http://arxiv.org/abs/1907.11692
Acknowledgements
Not applicable.
Funding
NCI R00CA245899, Doris Duke Charitable Foundation 2020080, DOD Grant (W81XWH-21-PCRP-DSA), DOD CDMRP award (HT9425-23-1-0023), Mark Foundation Emerging Leader Award, PCF-Movember Challenge Award.
Author information
Authors and Affiliations
Contributions
Conceptualization: HAE, KLK, and EMV; Methodology: HAE, KLK, and EMV; Software: HAE, PST, and VMA; Investigation: HAE, PST, VMA, IBR, DS, EMV, and KLK; Writing, original draft: KLK; Writing, review and editing: HAE, PST, VMA, IBR, DS, EMV, and KLK; Visualization: HAE, and IBR; Supervision: KLK and EMV; Funding acquisition: HAE, KLK and EMV.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The data for this analysis were derived from the EHRs of patients with lung cancer who had genomic profiling performed through the Dana-Farber Cancer Institute (DFCI) PROFILE [27] precision medicine effort or as a standard of care clinical test from June 26, 2013, to July 2, 2018. PROFILE participants consented to medical records review and genomic profiling of their tumor tissue. PROFILE was approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board (protocol #11-104 and #17-000); this supplemental retrospective analysis was declared exempt from review, and informed consent was waived for the standard of care genoty** patients given the minimal risk of data analysis, also by the Dana-Farber/Harvard Cancer Center Institutional Review Board (protocol #16-360). All methods were performed in accordance with the Declaration of Helsinki and approved by the Institutional Review Board at Dana Farber Cancer Institute.
Consent for publication
Not applicable.
Competing interests
Dr. Kehl reports serving as a consultant/advisor to Aetion, receiving funding from the American Association for Cancer Research related to this work, and receiving honoraria from Roche and IBM. Dr. Schrag reports compensation from JAMA for serving as an Associate Editor and from Pfizer for giving a talk at a symposium. She has received research funding from the American Association for Cancer Research related to this work and research funding from GRAIL for serving as the site-PI of a clinical trial. Unrelated to this work, Dr. Van Allen reports serving in advisory/consulting roles to Tango Therapeutics, Genome Medical, Invitae, Enara Bio, Janssen, Manifold Bio, and Monte Rosa; receiving research support from Novartis and BMS; holding equity in Tango Therapeutics, Genome Medical, Syapse, Enara Bio, Manifold Bio, Microsoft, and Monte Rosa; and receiving travel reimbursement from Roche/Genentech. Pavel Trukhanov reports ownership interest in NoRD Bio. The remaining authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Supplementary Figures.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Elmarakeby, H.A., Trukhanov, P.S., Arroyo, V.M. et al. Empirical evaluation of language modeling to ascertain cancer outcomes from clinical text reports. BMC Bioinformatics 24, 328 (2023). https://doi.org/10.1186/s12859-023-05439-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12859-023-05439-1