Abstract
Background
Several case–control studies have been conducted to clarify the association between the tumor necrosis factor alpha (TNF-α) -G308A polymorphism and risk of osteoarthritis (OA); however, the results are inconsistent. This meta-analysis was performed to clarify this issue using all the available evidence.
Methods
Eligible articles were retrieved by searching PubMed, Web of science and Google scholar. The strength of the association between the TNF-α -G308A polymorphism and risk of OA was assessed by odds ratios (ORs) with the corresponding 95% confidence interval (CI) for each study.
Results
Seven studies were included in the meta-analysis, which included 983 OA cases and 1355 controls. The pooled analysis based on all included studies showed a significantly increased OA risk in the recessive genetic model analysis (OR = 11.08, 95% CI = 4.75-25.86, p < 0.001) and in the A allele vs. G allele analysis (OR = 2.30, 95% CI = 1.08-4.90). However, there was no statistical difference in the dominant genetic model analysis (OR = 2.45, 95% CI = 0.95-6.27, p = 0.06). Furthermore, we found that OA patients had a higher frequency of the AA genotype (OR = 10.49, 95% CI = 4.47-24.61) and GA genotype (OR = 1.78, 95% CI = 1.03-3.08) compared with the control population.
Conclusion
Our results suggested that the TNF-α -G308A polymorphism were associated with an increased risk of OA.
Similar content being viewed by others
Background
As one of the most common age-related disease leading to restrictions in daily activities of the elderly, osteoarthritis (OA) is set to become the 4th-highest impact condition in women and the 8th-highest in men. The prevalence of OA is steadily increasing across all age groups [1]. Given the high morbidity and huge economic burden of OA, there is an urgent need to define the pathogenic factors involved in OA development. OA is a multistep, multifactorial disease that involves a complex interplay between genetic and environmental factors. Besides the traditional common risk factors, such as aging, obesity, previous injury, smoking habit and hormone therapy [2], recent studies have revealed that inflammatory processes play a pivotal role in OA pathogenesis [3]. Proinflammatory cytokines are now implicated as important mediators in OA, and TNF-α and interleukin 1 beta (IL-1β) are considered the major factors.
TNF-α is a multifunctional pro-inflammatory cytokine involved in various physiological and pathological processes, including inflammation, immunoregulation, proliferation and apoptosis [4]. TNF-α is produced by chondrocytes, mononuclear cells, osteoblasts and synovial tissues, and can stimulate its own production and induce chondrocytes and synovial cells to produce other cytokines. TNF-α also induces osteoclastic bone resorption [5] and destruction of cartilages [6]. Elevated levels of TNF-α have been found in the synovial fluid, synovial membrane, subchondral bone and cartilage of OA patients [7, 8], confirming its important roles in OA pathogenesis.
Genetic variations within the TNF-α promoter could alter the transcription and translation of TNF-α [9]. Polymorphisms in the promoter region of TNF-α have been found at -G238A, -G308A, -C863A, and -C857A, among which, the -G308A polymorphism is the most commonly studied. Previous studies found that the expression of TNF-α is higher in patients carrying the -308A allele than in those carrying the -308G allele, suggesting that this polymorphism has functional implications in inflammation and may be associated with inflammatory diseases [10]. In fact, previous studies have demonstrated the association of this polymorphism with certain inflammatory diseases, such as fatty liver disease [11], hepatitis B virus infection [12], systemic lupus erythematosus [13], inflammatory bowel disease [14], arteritis [15] and rheumatoid arthritis [16]. Several case–control studies have been conducted to clarify the association between the TNF-α -G308A polymorphism and risk of OA [17–23]; however, the results are inconsistent. Han’s study [20] and Ji’s study [22] found that the frequency of A allele was significantly higher in the OA cases; however, Sezgin’s study [19] and Valle’s study [21] found no association between the TNF-α -G308A polymorphism and OA risk. In view of the uncertain association between TNF-α -G308A polymorphism and OA risk, we sought to obtain more precise information by conducting a meta-analysis including all of the evidence produced to date.
Methods
Search strategy
Eligible articles were retrieved by searching the PubMed bibliographical database, Web of science and Google scholar (up to January 16, 2014) using the following combination of keywords: (tumour necrosis factor-alpha) OR (tumour necrosis factor-α) OR (TNF-α)); OA OR osteoarthritis; polymorphism OR polymorphisms OR variants OR variant. In addition, we checked the references in reviews and in the retrieved articles to avoid missing any relevant studies. There was no restriction on language in the search. The article selection was carried out by both YW and SK independently to minimize bias and human error.
Inclusion and exclusion criteria
To be included in this meta-analysis, an article had to provide the following information: 1) the number of OA cases and controls; and 2) the number of individuals with GG, GA and AA in both OA cases and controls. Those not designed as case–control studies, systemic reviews, and those that provided no usable data were excluded.
Data extraction
Two independent reviewers used a predesigned data extraction table to extract the data. The following information was extracted from each included article: journal name, first author, publication year, ethnicity and country, OA phenotype, source of controls, the number of genotypes in OA cases and controls, and the results of the study. Disagreement was resolved by discussion.
Statistical analysis
The strength of the association between the TNF-α -G308A polymorphism and risk of OA was assessed by odds ratios (ORs) with the corresponding 95% confidence interval (CI) for each study. The ORs and their 95% CIs were assessed for the following genetic models: 1) the dominant genetic model (AA + GA vs. GG) (A stands for the minor allele and G stands for the major allele); 2) the recessive genetic model (AA vs. GA + GG) and 3) the allele A vs. allele G analysis. The genotype frequencies of GG, GA and, AA were also assessed using the same method. A chi-squared (χ2) test was used to assess heterogeneity across studies. A fixed effect model was used when no heterogeneity existed among the studies. Otherwise, a random effect model was used. Meta-regression analysis was performed to find any sources of heterogeneity. Subgroup analysis for ethnicity (Asian and Caucasian), design type (HCC (hospital based case–control study) and PCC (population based case–control study)) and sample size (smaller (total sample <300) and larger (total sample ≥300)) was conducted. Potential publication bias was assessed using a funnel plot, and calculated using Begg’s and Egger’s tests (P < 0.05 was considered as significant publication bias). Influence analysis was performed by omitting each study to find potential outliers. In the control populations, the Hardy-Weinberg equilibrium (HWE) was tested using a chi- squared (χ2) test. Two authors performed the statistical analysis independently and obtained the same results. STATA software (version 11; Stata Corporation, College Station, TX, USA) was used to perform the statistical analyses. P values less than 0.05 were considered statistically significant.
Results
Literature selection and study characteristics
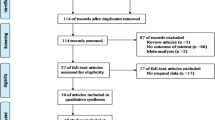
A total of 506 articles were retrieved from PubMed, Web of science and Google Scholar, 492 of which were excluded after screening the titles and abstracts (469 were irrelevant studies and 23 were duplicate studies). Fourteen articles were selected for detailed assessment and seven of them were excluded (three were not about G308A, three were not about OA risk and one had no usable data). Finally, seven studies were included in this meta-analysis, which included 983 OA cases and 1355 controls. The detailed selection procedure is shown in Figure 1. Genotype distributions in the controls of Moos’s study [17] were not in agreement with HWE. Most of the cases were knee OA, only one study recruited some hip OA cases [17]. The detailed characteristics of the included studies are shown in Table 1.
Quantitative data synthesis
The pooled analysis based on all included studies showed a significantly increased risk in the recessive genetic model analysis (OR = 11.08, 95% CI = 4.75-25.86, p < 0.001) (Figure 2B) and in the A allele vs. G allele analysis (OR = 2.30, 95% CI = 1.08-4.90, p < 0.001) (Figure 2C). However, there was statistically non-significant tendency towards decreased risk in the dominant genetic model analysis (OR = 2.45, 95% CI = 0.95-6.27, p = 0.06) (Figure 2A).We then assessed the genotype frequencies based on all included studies. The results showed that OA patients had a higher frequency of the AA genotype (OR = 10.49, 95% CI = 4.47-24.61, p = 0.001) (Figure 3A) and GA genotype (OR = 1.78, 95% CI = 1.03 -3.08, p = 0.006) (Figure 3B) compared with the control population. However, there was no significant difference for the GG genotype (OR = 0.77, 95% CI = 0.53-1.12, p = 0.14) (Figure 3C). The detailed results are shown in Table 2.
Tests of heterogeneity and subgroup analysis
We found heterogeneities in the dominant genetic model analysis (p < 0.01). A random effects model was adopted in these two analyses. Furthermore, a meta-regression analysis was conducted to find potential sources of heterogeneities. Unfortunately, the common variables, such as publication year, ethnicity, study design type and sample size were not significant sources of heterogeneities. However, we still performed subgroup analysis based on ethnicity, study design type (HCC or PCC,) and total sample size, because such subgroup analysis was valuable. The detailed results are shown in Table 2.
Sensitivity analysis
Influence analysis was performed to assess the sensitivity of each individual study on the pooled ORs, by sequential omission of each individual study. The results suggested that Han’s study [20] significantly affected almost all of the pooled ORs (Figure 4A-C). Therefore, we calculated the results again with Han’s study [20] omitted. The results are shown in Table 3 and Additional file 1: Figure S1; Additional file 2: Figure S2. The pooled analysis showed an increased risk in all three analyses: the dominant model analysis (OR = 1.72, 95% CI = 1.35 -2.20, p < 0.001), the recessive model analysis (OR = 4.84, 95% CI = 1.90 -12.35, p = 0.001) and A allele vs. G allele analysis (OR = 1.75, 95% CI = 1.40 -2.19, p < 0.001) (Additional file 1: Figure S1A-C). We also re-assessed the genotype frequency of GG, GA and AA; however, the results were almost the same as those including all the studies (Additional file 2: Figure S2A-C).
Influence analysis for TNF-α -308 polymorphism in the overall analysis: (A) dominant model analysis; (B) recessive model analysis; (C) A allele vs. G allele analysis. These figures show the influence of individual studies on the pooled ORs. The middle vertical axis represents the overall ORs and the two vertical axes represent its 95% CIs. Open circles represent the pooled OR when the selected study is omitted in this meta-analysis. The two ends of the dotted lines indicate the 95% CI.
Publication bias
Funnel plots were used to estimate qualitatively the potential publication bias. Taking the the recessive model analysis and A allele vs. G allele analysis as examples, the shapes of the funnel plots did not indicate any obvious asymmetry (Figure 5A,B). Furthermore, Begg’s and Egger’s tests were used to estimate quantitatively the potential publication bias. The p values were all greater than 0.05, indicating no publication bias.
Discussion
To the best of our knowledge, this is the first meta-analysis to evaluate the association between the TNF-α -308 polymorphism and risk of OA. In this meta-analysis, we discovered an increased OA risk in the recessive genetic model analysis and in the A allele vs. G allele analysis. Furthermore, we found a higher frequency of the AA and GA genotype in OA patients.
TNF-α is a potent pro-inflammatory cytokine in inflammatory diseases and the immune response. Higher levels of TNF-α were found in OA patients compared with the control population [24]. TNF-α can induce the production of other cytokines, matrix metalloproteinases and prostaglandins 9, and inhibit the synthesis of proteoglycans and type II collagen; thus it plays a pivotal role in cartilage matrix degradation and bone resorption in OA [25, 26]. Single nucleotide conversion from guanine (G) to adenine (A) at position -308 is the most common polymorphism in general populations and this transition has been shown to influence the expression of TNF-α. According to previous study, the -308 allele A is associated with about 6 fold increased transcriptional activity and higher protein levels of TNF-α [27]. Valle’s study [21] also found that OA patients with -308 GG genotype expressed less TNF-α mRNA than patients with the GA genotype. Therefore, the TNF-α -308 polymorphism may have some effect on the hosts’ susceptibility to OA by altering TNF-α expression. In support of this, our meta-analysis found an increased risk of OA in the AA vs. GG analysis and the recessive model analysis, and higher frequencies of the AA and GA genotypes in OA patients. However, by subgroup analysis, we found that the relationship of TNF-α-308 polymorphism and OA risk only existed among Asian population but not among Caucasion population. In recent GWAS studies, 11 OA susceptibility loci with genome-wide significance levels was indentified. Among them, a region containing HLA class II/III genes, which close to the region habouring TNF gene, showed association in Asians but not in Europeans [28–30], other nine loci reached genome-wide significance in Europeans[31]. These findings exactly explained why the relationship of TNF-α-308 polymorphism and OA risk only existed among Asian population but not among Caucasion population.
Influence analysis suggested that Han’s study [20] might have significantly affected the results; however, when omitting Han’s study, the conclusions showed good consistency with the previous results. Therefore, the conclusions from our meta- analysis are sound and reliable.
Recently, we have noticed that some studies have been conducted to explore the efficacy of anti-TNF therapy on OA. Güler-Yüksel’s study have found that treatment with TNF-α inhibitor might reduce hand OA in patients with rheumatoid arthritis[32], which shed light to the role of anti-TNF-α in treatment of hand OA. However, a recently published RCT failed to confirm the efficacy of anti-TNF-α in treatment of primary hand OA [33], and the author speculated that TNF α is not the right target in order to improve symptoms in hand OA and TNF α blockers may exert a differential effect on pain and structure. However, we just have different opinions. We suppose that anti-TNF-α treatment may be more efficient in patients with -308 allele A genotype because the -308 allele A is associated with about 6 fold increased transcriptional activity and higher protein levels of TNF-α. Therefore, the detection of -308 polymorphism of TNF-α is necessary before the treatment of TNF-α blocker. Further studies are needed to confirm our speculation.
Although the primary results of our meta-analysis are suggestive, some limitations may still exist. Firstly, there was heterogeneity between studies of the -308 polymorphism and the heterogeneity may have distorted the meta-analysis. Unfortunately, meta-regression analysis failed to find the potential heterogeneity. In addition, no quality tool has been used to assess the risk of bias in the included studies, study quality could be one factor missing from the heterogeneity analysis. Secondly, the number of OA cases and studies included in the Caucasian-specific meta-analysis was low. This study may not have enough power to explore the association between the TNF-α -308 polymorphism and risk of OA in the Caucasian population. Thirdly, the results of the meta-analysis were based on unadjusted estimates owing to the lack of adjusted estimates. Currently, some risk factors have been identified, such as aging, obesity, previous injury, smoking habit and hormone therapy. A more precise analysis could be performed if these data were available. Finally, gene-gene interactions were not fully addressed in the meta-analysis because of the lack of relevant data. In fact, the TNF-α gene is located close to the human leucocyte antigen (HLA) class II/III region, in which some SNPs, such as rs7775228 and rs10947262, have been identified as associated with risk of knee OA [30, 34]. Therefore, the positive results from our meta-analysis may result from the linkage disequilibrium of the proximal susceptible SNPs. Further studies on the all SNPs in this region and OA risk are needed.
Conclusions
In conclusion, this comprehensive meta-analysis has evaluated all published data currently available on the TNF-α -308 polymorphism and risk of OA. Our meta-analysis suggested that the AA and GA genotypes might increase the risk of OA compared with the GG genotype, which may be explained by the higher expression of TNF-α in the -308A allele carriers than in the -308G carriers.
Abbreviations
- TNF-α:
-
Tumor necrosis factor alpha
- OA:
-
Osteoarthritis
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- IL-1β:
-
Interleukin 1 beta
- HCC:
-
Hospital based case–control study
- PCC:
-
Population based case–control study
- HWE:
-
Hardy-Weinberg equilibrium
- HLA:
-
Human leucocyte antigen.
References
Murray CJL, Lopez AD: The Global Burden of Disease: A Comprehensive Assessment of Mortality and Disability from Diseases, Injuries, and Risk Factors in 1990 and Projected to 2020. Cambridge: Harvard School of Public Health, on behalf of the World Health Organization; Global Burden of Disease and Injury Series, 1996. Vol. 1
Prasanna SS, Korner-Bitensky N, Ahmed S: Why do people delay accessing health care for knee osteoarthritis? Exploring beliefs of health professionals and lay people. Physiother Can. 2013, 65 (1): 56-63. 10.3138/ptc.2011-50.
Berenbaum F, Eymard F, Houard X: Osteoarthritis, inflammation and obesity. Curr Opin Rheumatol. 2013, 25 (1): 114-118. 10.1097/BOR.0b013e32835a9414.
Esposito E, Cuzzocrea S: TNF-alpha as a therapeutic target in inflammatory diseases, ischemia-reperfusion injury and trauma. Curr Med Chem. 2009, 16 (24): 3152-3167. 10.2174/092986709788803024.
Pelletier JP, Martel-Pelletier J, Abramson SB: Osteoarthritis, an inflammatory disease: potential implication for the selection of new therapeutic targets. Arthritis Rheum. 2001, 44 (6): 1237-1247. 10.1002/1529-0131(200106)44:6<1237::AID-ART214>3.0.CO;2-F.
Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H: Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011, 7 (1): 33-42. 10.1038/nrrheum.2010.196.
Stannus O, Jones G, Cicuttini F, Parameswaran V, Quinn S, Burgess J, Ding C: Circulating levels of IL-6 and TNF-α are associated with knee radiographic osteoarthritis and knee cartilage loss in older adults. Osteoarthritis Cartilage. 2010, 18 (11): 1441-1447. 10.1016/j.joca.2010.08.016.
Xue J, Wang J, Liu Q, Luo A: Tumor necrosis factor-α induces ADAMTS-4 expression in human osteoarthritis chondrocytes. Mol Med Rep. 2013, 8 (6): 1755-1760.
Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW: Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A. 1997, 94 (7): 3195-3199. 10.1073/pnas.94.7.3195.
Messer G, Spengler U, Jung MC, Honold G, Blömer K, Pape GR, Riethmüller G, Weiss EH: Polymorphic structure of the tumor necrosis factor (TNF) locus: an NcoI polymorphism in the first intron of the human TNF-beta gene correlates with a variant amino acid in position 26 and a reduced level of TNF-beta production. J Exp Med. 1991, 173 (1): 209-219. 10.1084/jem.173.1.209.
Aller R, de Luis DA, Izaola O, González Sagrado M, Conde R, Alvarez Gago T, Pacheco D, González JM, Velasco MC: G308A polymorphism of TNF-alpha gene is associated with insulin resistance and histological changes in non alcoholic fatty liver disease patients. Ann Hepatol. 2010, 9 (4): 439-444.
Zhang TC, Zhao YQ, Hu GL, Liu XQ, Huang XK: The relationship between tumour necrosis factor-α gene polymorphism and susceptibility and clearance of the persistent hepatitis B virus infection in a Chinese population: a meta-analysis. Clin Microbiol Infect. 2013, doi:10.1111/1469-0691.12257
Ahmed HH, Taha FM, Darweesh HE, Morsi HM: Association between TNF promoter -308 G>A and LTA 252 A>G polymorphisms and systemic lupus erythematosus. Mol Biol Rep. 2014, 41 (4): 2029-2036. 10.1007/s11033-014-3051-7.
Tong Q, Zhao L, Qian XD, Zhang LL, Xu X, Dai SM, Cai Q, Zhao DB: Association of TNF-α polymorphism with prediction of response to TNF blockers in spondyloarthritis and inflammatory bowel disease: a meta-analysis. Pharmacogenomics. 2013, 14 (14): 1691-1700. 10.2217/pgs.13.146.
Sandhya P, Danda S, Danda D, Lonarkar S, Luke SS, Sinha S, Joseph G: Tumour necrosis factor (TNF)-α-308 gene polymorphism in Indian patients with Takayasu’s arteritis - a pilot study. Indian J Med Res. 2013, 137 (4): 749-752.
Maxwell JR, Potter C, Hyrich KL, Barton A, Worthington J, Isaacs JD, Morgan AW, Wilson AG, Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate: Association of the tumour necrosis factor-308 variant with differential response to anti-TNF agents in the treatment of rheumatoid arthritis. Hum Mol Genet. 2008, 17 (22): 3532-3538. 10.1093/hmg/ddn245.
Moos V, Rudwaleit M, Herzog V, Höhlig K, Sieper J, Müller B: Association of genotypes affecting the expression of interleukin-1beta or interleukin-1 receptor antagonist with osteoarthritis. Arthritis Rheum. 2000, 43 (11): 2417-2422. 10.1002/1529-0131(200011)43:11<2417::AID-ANR7>3.0.CO;2-R.
Muñoz-Valle JF, Martin-Márquez BT: -308 tumor necrosis factor-alpha promoter polymorphism in patients with rheumatoid arthritis and osteoarthritis in the west of mexico: preliminary results(j). Rev Vaccimonitor (Vacunologíay Temasafines). 2002, 11 (4): 4-
Sezgin M, Barlas IO, Ankarali HC, Altintaş ZM, Türkmen E, Gökdoğan T, Sahin G, Erdal ME: Tumour necrosis factor alpha -308G/A gene polymorphism: lack of association with knee osteoarthritis in a Turkish population. Clin Exp Rheumatol. 2008, 26 (5): 763-768.
Han L, Song JH, Yoon JH, Park YG, Lee SW, Choi YJ, Nam SW, Lee JY, Park WS: TNF-α and TNF-β Polymorphisms are Associated with Susceptibility to Osteoarthritis in a Korean Population. Korean J Pathol. 2012, 46 (1): 30-37. 10.4132/KoreanJPathol.2012.46.1.30.
Muñoz-Valle JF, Oregón-Romero E, Rangel-Villalobos H, Martínez-Bonilla GE, Castañeda-Saucedo E, Salgado-Goytia L, Leyva-Vázquez MA, Illades-Aguiar B, Alarcón-Romero Ldel C, Espinoza-Rojo M, Parra-Rojas I: High expression of TNF alpha is associated with -308 and -238 TNF alpha polymorphisms in knee osteoarthritis. Clin Exp Med. 2012, Epub ahead of print
Ji B, Shi J, Cheng X, Zhou J, Zhou Q, Cao C, Pang J: Association analysis of two candidate polymorphisms in the tumour necrosis factor-α gene with osteoarthritis in a Chinese population. Int Orthop. 2013, 37 (10): 2061-2063. 10.1007/s00264-013-1931-4.
Cheng XY, Cao CF, Ji B: Association of tumor necrosis factor- alpha -308 polymorphism with osteoarthritis in a Chinese population. Chin J Orthop Trauma. 2013, 15 (7): 607-609.
Amin AR: Regulation of tumor necrosis factor-alpha and tumor necrosis factor converting enzyme in human osteoarthritis. Osteoarthritis Cartilage. 1999, 7: 392e4-
Goldring SR, Goldring MB: The role of cytokines in cartilage matrix degeneration in osteoarthritis. Clin Orthop Relat Res. 2004, 427 (Suppl): S27-S36.
Klooster AR, Bernier SM: Tumor necrosis factor alpha and epidermal growth factor act additively to inhibit matrix gene expression by chondrocyte. Arthritis Res Ther. 2005, 7 (1): R127-R138. 10.1186/ar1464.
Zhuang L, Ma W, Cai D, Zhong H, Sun Q: Associations between tumor necrosis factor-α polymorphisms and risk of psoriasis: a meta-analysis. PLoS One. 2013, 8 (12): e68827-10.1371/journal.pone.0068827.
Miyamoto Y, Shi D, Nakajima M, Ozaki K, Sudo A, Kotani A, Uchida A, Tanaka T, Fukui N, Tsunoda T, Takahashi A, Nakamura Y, Jiang Q, Ikegawa S: Common variants in DVWA on chromosome 3p24.3 are associated with susceptibility to knee osteoarthritis. Nat Genet. 2008, 40 (8): 994-998. 10.1038/ng.176.
Meulenbelt I, Chapman K, Dieguez-Gonzalez R, Shi D, Tsezou A, Dai J, Malizos KN, Kloppenburg M, Carr A, Nakajima M, van der Breggen R, Lakenberg N, Gomez-Reino JJ, Jiang Q, Ikegawa S, Gonzalez A, Loughlin J, Slagboom EP: Large replication study and meta-analyses of DVWA as an osteoarthritis susceptibility locus in European and Asian populations. Hum Mol Genet. 2009, 18 (8): 1518-1523. 10.1093/hmg/ddp053.
Nakajima M, Takahashi A, Kou I, Rodriguez-Fontenla C, Gomez-Reino JJ, Furuichi T, Dai J, Sudo A, Uchida A, Fukui N, Kubo M, Kamatani N, Tsunoda T, Malizos KN, Tsezou A, Gonzalez A, Nakamura Y, Ikegawa S: New sequence variants in HLA class II/III region associated with susceptibility to knee osteoarthritis identified by genome-wide association study. PLoS One. 2010, 5 (3): e9723-10.1371/journal.pone.0009723.
Rodriguez-Fontenla C, Calaza M, Evangelou E, Valdes AM, Arden N, Blanco FJ, Carr A, Chapman K, Deloukas P, Doherty M, Esko T, Garcés Aletá CM, Gomez-Reino Carnota JJ, Helgadottir H, Hofman A, Jonsdottir I, Kerkhof HJ, Kloppenburg M, McCaskie A, Ntzani EE, Ollier WE, Oreiro N, Panoutsopoulou K, Ralston SH, Ramos YF, Riancho JA, Rivadeneira F, Slagboom PE, Styrkarsdottir U, Thorsteinsdottir U, et al: Assessment of osteoarthritis candidate genes in a meta-analysis of nine genome-wide association studies. Arthritis Rheumatol. 2014, 66 (4): 940-949. 10.1002/art.38300.
Güler-Yüksel M, Allaart CF, Watt I, Goekoop-Ruiterman YP, de Vries-Bouwstra JK, van Schaardenburg D, van Krugten MV, Dijkmans BA, Huizinga TW, Lems WF, Kloppenburg M: Treatment with TNF-α inhibitor infliximab might reduce hand osteoarthritis in patients with rheumatoid arthritis. Osteoarthritis Cartilage. 2010, 18 (10): 1256-1262. 10.1016/j.joca.2010.07.011.
Chevalier X, Ravaud P, Maheu E, Baron G, Rialland A, Vergnaud P, Roux C, Maugars Y, Mulleman D, Lukas C, Wendling D, Lafforgue P, Loeuille D, Foltz V, Richette P, On behalf of the French section of osteoarthritis: Adalimumab in patients with hand osteoarthritis refractory to analgesics and NSAIDs: a randomised, multicentre, double-blind, placebo-controlled trial. Ann Rheum Dis. 2014, doi:10.1136/annrheumdis-2014-205348
Shi D, Zheng Q, Chen D, Zhu L, Qin A, Fan J, Liao J, Xu Z, Lin Z, Norman P, Xu J, Nakamura T, Dai K, Zheng M, Jiang Q: Association of single-nucleotide polymorphisms in HLA class II/III region with knee osteoarthritis. Osteoarthritis Cartilage. 2010, 18 (11): 1454-1457. 10.1016/j.joca.2010.07.009.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2474/15/373/prepub
Acknowledgements
This work was funded by the Action Plan (major research) projects for three years by the development of Chinese medicine in Shanghai (No.ZYSNXD-CC-ZDYJ033) and Shanghai Science and Technology 2012 “Technology Innovation Action Plan” project funding (No.12401904700).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
All authors declare that they have no competing interest.
Authors’ contributions
YW conceived of the study, both YW and SK participated in the design of the study and performed the statistical analysis and draft the manuscript. Both authors read and approved the final manuscript.
Electronic supplementary material
12891_2014_2359_MOESM1_ESM.tiff
Additional file 1: Figure S1: Meta-analysis of TNF-α -308 polymorphism and OA risk after omitting Han’s study: (A) dominant genetic model analysis; (B) recessive genetic model analysis; (C) A allele vs. G allele analysis. (TIFF 12 MB)
12891_2014_2359_MOESM2_ESM.tiff
Additional file 2: Figure S2: Meta-analysis of TNF-α -308 genotypes and OA risk after omitting Han’s study: (A) AA genotype frequency; (B) GA genotype frequency; (C) GG genotype frequency. (TIFF 12 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Kou, S., Wu, Y. Meta-analysis of tumor necrosis factor alpha -308 polymorphism and knee osteoarthritis risk. BMC Musculoskelet Disord 15, 373 (2014). https://doi.org/10.1186/1471-2474-15-373
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2474-15-373