Abstract
Background
Liver regeneration (LR) after 2/3 partial hepatectomy (PH) is one of the most studied models of cell, organ, and tissue regeneration. Although the transcriptional profile analysis of regenerating liver has been carried out by many reserachers, the dynamic protein expression profile during LR has been rarely reported up to date. Therefore, this study aims to detect the global proteomic profile of the regenerating rat liver following 2/3 hepatectomy, thereby gaining some insights into hepatic regeneration mechanism.
Results
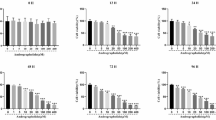
Protein samples extracted from the sham-operated and the regenerating rat livers at 6, 12, 24, 72, 120 and 168 h after PH were separated by IEF/SDS-PAGE and then analyzed by MALDI-TOF/TOF mass spectrometry. Compared to sham-operated groups, there were totally 220 differentially expressed proteins (including 156 up-regulated, 62 down-regulated, and 2 up/down-regulated ones) identified in the regenerating rat livers, and most of them have not been previously related to liver regeneration. According to the expression pattern analysis combined with gene functional analysis, it showed that lipid and carbohydrate metabolism were enhanced at the early phase of LR and continue throughout the regeneration process. Ingenuity Pathway Analysis indicated that YWHAE protein (one of members of the 14-3-3 protein family) was located at the center of pathway networks at all the timepoints after 2/3 hepatectomy under our experimental conditions, maybe suggesting a central role of this protein in regulating liver regeneration. Additionally, we also revealed the role of Cdc42 (cell division cycle 42) in the termination of LR.
Conclusions
For the first time, our proteomic analysis suggested an important role of YWHAE and pathway mediated by this protein in liver regeneration, which might be helpful in expanding our understanding of LR amd unraveling the mechanisms of LR.
Similar content being viewed by others
Background
The liver is one of the vital organs of the body with mutiple important responsibilities, including metabolism, maintenance of water balance, bile acid production and excretion, detoxification, immune response and so on [1]. In addition, the liver becomes distinguished from other organs mainly by its amazing regenerative ability, which is primarily attributable to the quick reentry of highly differentiated quiescent hepatic cells into the cell cycle in response to liver injury induced by surgical resection (e.g., surgerical resection, pathogenic or chemical factors [2]. Among various liver injury models, partial hepatectomy (PH), resulting in the loss of approximately 70% of the liver volume, is now widely utilized for studying liver regeneration in experimental animals. According to previous reports, many cytokines (e.g., TNF-α, IL-6) are upregulated within 0-6 h after 70% liver resection. DNA synthesis begins at 12-16 h following PH, peaking around at 24-48 h, which results in a steep increase of liver mass at 72 h after PH, followed by the rough restoration of liver mass to normal around at 168 h after PH [3]. In general, rat liver regeneration lasts about 7 days, during which involves many biological events including cell activation, cell differentiation, proliferation and its regulation, redifferentiation, reestablishment of liver structure and function as well, inferring that the mechanism underlying this process is highly complex [4]. Therefore, in order to understand clarify the regenerative process of the liver, it is very essential to clarify the overall and systematically molecular basis of this stringently regulated process.
So far, the high-throughput analysis technologies, such as genechip, serial analysis of gene expression (SAGE), have been extensively used by researchers to identify transcriptome profiles of the regenerating livers, and obtained the valuable data for understanding mechanism of LR. Especially in 2009, Wang et al. analyzed the gene expression profiling of regenerating rat livers at different recovery time points after PH, finding that there are totally 1004 known genes and 857 unknown genes associated with LR [5]. However, the data from microarray analysis is too limited to quantitatively analyze protein levels, or even to reflect the final biological effect of genes. So, despite that the gene expression pattern has come under intense scrutiny, a differential proteomic study could better help to elucidate how the process is triggered and regulated.
Recently, the emergence of comparative proteomics technologies, such as two-dimensional electrophoresis (2-DE), high performance liquid chromatography (HPLC), mass spectrometry (MS) and mass fingerprinting, provides a promising approach for elucidating the mechanism of LR [6]. As a protein separation technique, 2-DE has been widely employed in separating and quantifying proteins that are differentially expressed in proteomics research. Depending on this method, the proteins whose levels were significantly changed under a defined physiological condition can be screened out and identified. For instance, Strey and his coworkers applied two-dimensional gel electrophoresis approach to measure protein expression changes in mouse livers at 6 h and 12 h after PH, and identified twelve up-regulated (at least 2-fold) proteins related to signaling and metabolic pathways [7]. Nevertheless, this technology has some striking limitations such as low load ability, poor separation of hydrophobic, acidic and alkaline proteins. And HPLC or MS techniques overcome the shortcomings of 2-DE, and suit differential proteomics study much better. Also, other techniques such as multi-dimension chromatogram, protein chips have been utilized to complement or substitute this conventional method. Presently, the method of 2-DE separation combined with MS identification is now in common use in this research field. For example, He et al. introduced this method to detect protein expression profiles in rat regenerating livers at 1 hour after PH, and identified a total of 24 differentially expressed proteins. In addition, He’s research team applied 2-DE in combination with MALDI-QTOF-MS compared protein expression patterns between sham-operation group and 7-hour hepatectomized group to identify proteins whose expression may be altered after PH, and discovered 29 differentially expressed proteins [Pathway analysis Pathway network analysis of the identified proteins differentially expressed at the 6-, 12-, 24-, 72-, 120- and 168-h time points were carried out with the Shortest Paths and the Analyze Network options of Ingenuity Pathway Analysis 9.0 software, a web-delivered application (http://www.Ingenuity.com) that can identify major biological themes during LR and predict the key networks functioning at different phases of regeneration process. The Shortest Paths option can incoporate all given proteins into the shortest possible paths. The network is built dynamically, connecting all the proteins through direct mechanistic interactions on the basis of manually curated publications.
References
Su LJ, Ding GW, Yang ZL, Zhang SB, Yang YX, Xu CS: Expression patterns and action analysis of genes associated with hepatitis virus infection during rat liver regeneration. World J Gastroenterol 2006, 12: 7626-7634.
Fan BL, Zhang WL, Du MH, Zhu WL: Differential expression of the polo-like kinase 1 (plk1) gene during liver regeneration and hepatic oval cell proliferation. Zhonghua Gan Zang Bing Za Zhi 2013, 21: 705-706.
Xu C, Chen X, Chang C, Wang G, Wang W, Zhang L, Zhu Q, Wang L, Zhang F: Transcriptome analysis of hepatocytes after partial hepatectomy in rats. Dev Genes Evol 2010, 220: 263-274. 10.1007/s00427-010-0345-1
Riehle KJ, Dan YY, Campbell JS, Fausto N: New concepts in liver regeneration. J Gastroenterol Hepatol 2011, 26(Suppl 1):203-212.
Wang WB, Fan JM, Zhang XL, Xu J, Yao W: Serial expression analysis of liver regeneration-related genes in rat regenerating liver. Mol Biotechnol 2009, 43: 221-231. 10.1007/s12033-009-9199-z
Feng C, Li JL, Cao R, Chen P: Differential Proteomic Analysis of Plasma Membrane in Rat Liver at 72 Hours after 2/3 Partial Hepatectomy. Chin J Biochem Mol Biol 2012, 5: 751-760.
Strey CW, Winters MS, Markiewski MM, Lambris JD: Partial hepatectomy induced liver proteome changes in mice. Proteomics 2005, 5: 318-325. 10.1002/pmic.200400913
Guo F, Nian H, Zhang H, Huang L, Tang Y, **ao X, He D: Proteomic analysis of the transition from quiescent to proliferating stages in rat liver hepatectomy model. Proteomics 2006, 6: 3075-3086. 10.1002/pmic.200500322
Hsieh HC, Chen YT, Li JM, Chou TY, Chang MF, Huang SC, Tseng TL, Liu CC, Chen SF: Protein profilings in mouse liver regeneration after partial hepatectomy using iTRAQ technology. J Proteome Res 2009, 8: 1004-1013. 10.1021/pr800696m
Michalopoulos GK, DeFrances MC: Liver regeneration. Science 1997, 276: 60-66. 10.1126/science.276.5309.60
Lee C, Oh JI, Park J, Choi JH, Bae EK, Lee HJ, Jung WJ, Lee DS, Ahn KS, Yoon SS: TNFα mediated IL-6 secretion is regulated by JAK/STAT pathway but not by MEK phosphorylation and AKT phosphorylation in U266 multiple myeloma cells. Biomed Res Int 2013, 2013: 580135.
Taub R: Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol 2004, 5: 836-847. 10.1038/nrm1489
Cao H, Yu J, Xu W, Jia X, Yang J, Pan Q, Zhang Q, Sheng G, Li J, Pan X, Wang Y, Li L: Proteomic analysis of regenerating mouse liver following 50% partial hepatectomy. Proteome Sci 2009, 7: 48. 10.1186/1477-5956-7-48
Li X, **ong L, **e C, Cao J, Deng H, Lin Y, Cao R, Li J, Chen P, Liang S: Proteomics analysis of plasma membrane from liver sinusoidal endothelial cells after partial hepatectomy by an improved two-dimensional electrophoresis. Mol Cell Biochem 2010, 344: 137-150. 10.1007/s11010-010-0537-z
Yuan H, Zhang H, Wu X, Zhang Z, Du D, Zhou W, Zhou S, Brakebusch C, Chen Z: Hepatocyte-specific deletion of Cdc42 results in delayed liver regeneration after partial hepatectomy in mice. Hepatology 2009, 49: 240-249. 10.1002/hep.22610
Milland J, Tsykin A, Thomas T, Aldred AR, Cole T, Schreiber G: Gene expression in regenerating and acute-phase rat liver. Am J Physiol 1990, 259: G340-G347.
Alexander MC, Lomanto M, Nasrin N, Ramaika C: Insulin stimulates glyceraldehyde-3-phosphate dehydrogenase gene expression through cis-acting DNA sequences. Proc Natl Acad Sci U S A 1988, 85: 5092-5096. 10.1073/pnas.85.14.5092
Matos JM, Witzmann FA, Cummings OW, Schmidt CM: A Pilot Study of Proteomic Profiles of Human Hepatocellular Carcinoma in the United States. J Surg Res 2009, 155: 237-243. 10.1016/j.jss.2008.06.008
Sun Y, Deng X, Li W, Yan Y, Wei H, Jiang Y, He F: Liver proteome analysis of adaptive response in rat immediately after partial hepatectomy. Proteomics 2007, 7: 4398-4407. 10.1002/pmic.200600913
des Portes V, Pinard JM, Billuart P, Vinet MC, Koulakoff A, Carrié A, Gelot A, Dupuis E, Motte J, Berwald-Netter Y, Catala M, Kahn A, Beldjord C, Chelly J: A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell 1998, 92: 51-61. 10.1016/S0092-8674(00)80898-3
Rudinskiy N, Kaneko YA, Beesen AA, Gokce O, Régulier E, Déglon N, Luthi-Carter R: Diminished hippocalcin expression in Huntington’s disease brain does not account for increased striatal neuron vulnerability as assessed in primary neurons. J Neurochem 2009, 111: 460-472. 10.1111/j.1471-4159.2009.06344.x
Crumm S, Cofan M, Juskeviciute E, Hoek JB: Adenine nucleotide changes in the remnant liver: An early signal for regeneration after partial hepatectomy. Hepatology 2008, 48: 898-908. 10.1002/hep.22421
Higgins GM, Anderson RM: Experimental pathology of the liver: restoration of the liver of the white rat following partial surgical removal. Arch Pathol 1931, 12: 186-202.
Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Rigetti PG: Blue Silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 2004, 25: 1327-1333. 10.1002/elps.200305844
Yu LR, Zeng R, Shao XX, Wang N, Xu YH, **a QC: Identification of differentially expressed proteins between human hepatoma and normal liver cell lines by two-dimensional electrophoresis and liquid chromatography-ion trap mass spectrometry. Electrophoresis 2000, 21: 3058-3068. 10.1002/1522-2683(20000801)21:14<3058::AID-ELPS3058>3.0.CO;2-U
Laemmli UK: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1977, 227: 680-686.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31401209) and National Basic Research 973 Pre-research Program of China (No. 2010CB534905).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
X-GC carried out a series of specific experimental operations in this study, including animal model preparations, 2-DE MALDI TOF/TOF MS analysis and Western blotting assay, and participated in the drafting of this manuscript. C-SX is responsible for the general design of study, the analysis of experimental data, and the drafting of final version. Finally, both authors read and approved the final manuscript.
Electronic supplementary material
40659_2014_70_MOESM1_ESM.xls
Additional file 1: Table S1: Protein expression profiling of both sham-operated groups (highlighted in red) and partial-hepatectomized groups (highlighted in blue) in six clusters. Note: The gray-colored bins represent the ≥2-fold up-regulation; the spot-marked bins represent ≥2-fold down-regulation; the colorless bins represent the insignificant expression. Asterisk denotes those proteins that have been identified to be LR-related in other studies. (XLS 84 KB)
40659_2014_70_MOESM2_ESM.jpeg
Additional file 2: Figure S1: Pathway analysis results showing that differentially expressed proteins at each time points after 2/3 PH in rats can be sorted into a specific biological pathway. The root nodes in six subnetworks are all connected to YWHAE directly or indirectly. Red and green proteins were identified as differentially upregulated and downregulated, respectively. Other gray denoted the proteins that were down-regulated only in SO sample or detected only in SO sample. Lines connecting the molecules indicate molecular relationships. Real lines indicate direct interactions and dashed lines indicate indirect interactions. (JPEG 192 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Chen, XG., Xu, CS. Proteomic analysis of the regenerating liver following 2/3 partial hepatectomy in rats. biol res 47, 59 (2014). https://doi.org/10.1186/0717-6287-47-59
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/0717-6287-47-59