Abstract
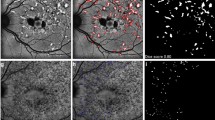
Retinitis pigmentosa is a retinal disorder leading to a progressive visual field loss and eventually to complete blindness, but an early diagnosis could delay its progression through specific therapies. Retinitis pigmentosa is characterized by typical pigment signs that accumulate in the different regions of the retina. Pigment signs could be detected by a low-cost diagnosis tool, analyzing visual fundus retinal images and applying segmentation algorithms to annotate the pigments, so that, in a telemedicine scenario, the segmented images could be forwarded to an ophthalmologist for a rapid diagnosis. Deep learning approaches might be appropriate for this problem, but they have rarely been used to address it. However, pigment segmentation is a challenging task due to image resolution, small size of pigments and their proximity with blood vessels with which they share similar colors, and inter-patient widely changing image features. Very recently, transformer architectures, based on the self-attention paradigm, have emerged in the deep learning community as a powerful yet not completely explored tool to learn features directly from the data. Nonetheless, they could not be directly exploited on small datasets, as they require a very large amount of data to learn meaningful features. To overcome the need for large training datasets, but also to reduce the high computation effort, hybrid architectures have been proposed, with the aim to combine the long-range relationship detection of transformers with the invariance and short-range detection properties of classical deep learning architectures. Here, we investigate the performances of the Group Transformer U-Net, a hybrid approach for pigment segmentation on fundus images. This hybrid architecture modifies the classical U-Net structure introducing bottleneck multihead self-attention blocks between convolutional layers in both the contracting and expanding paths of the network. We compare the results obtained with this approach with the ones of the standard U-Net, and we describe how these results are affected when using different loss functions for the learning process, or strategies to address class imbalance.




Similar content being viewed by others
REFERENCES
S. d’Ascoli, H. Touvron, M. Leavitt, A. Morcos, G. Biroli, and L. Sagun, “ConViT: Improving vision transformers with soft convolutional inductive biases,” Proc. Mach. Learn. Res. 139, 2286–2296 (2021). ar**v:2103.10697 [cs, stat]
S. A. Taghanaki, K. Abhishek, J. P. Cohen, J. Cohen-Adad, and G. Hamarneh, “Deep semantic segmentation of natural and medical images: A review,” Artif. Intell. Rev. 54, 137–178 (2021). https://doi.org/10.1007/s10462-020-09854-1
M. Berman, A. R. Triki, and M. B. Blaschko, “The Lovasz-Softmax Loss: A tractable surrogate for the optimization of the intersection-over-union measure in neural networks,” in IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Salt Lake City, Utah, 2018 (IEEE, 2018), pp. 4413–4421. https://doi.org/10.1109/CVPR.2018.00464
N. Brancati, M. Frucci, D. Gragnaniello, D. Riccio, V. Di Iorio, L. Di Perna, and F. Simonelli, “Learning-based approach to segment pigment signs in fundus images for Retinitis Pigmentosa analysis,” Neurocomputing 308, 159–171 (2018). https://doi.org/10.1016/j.neucom.2018.04.065
N. Brancati, M. Frucci, D. Riccio, L. D. Perna, and F. Simonelli, “Segmentation of pigment signs in fundus images for retinitis pigmentosa analysis by using deep learning,” in Image Analysis and Processing – ICIAP 2019, Ed. by E. Ricci, S. Rota Bulò, C. Snoek, O. Lanz, S. Messelodi, and N. Sebe, Lecture Notes in Computer Science, vol. 11752 (Springer, Cham, 2019), pp. 437–445. https://doi.org/10.1007/978-3-030-30645-8_40
T. Falk, D. Mai, R. Bensch, Ö. Çiçek, A. Abdulkadir, Y. Marrakchi, A. Böhm, J. Deubner, Z. Jäckel, K. Seiwald, A. Dovzhenko, O. Tietz, C. D. Bosco, S. Walsh, D. Saltukoglu, T. L. Tay, M. Prinz, K. Palme, M. Simons, I. Diester, T. Brox, and O. Ronneberger, “UрNet: Deep learning for cell counting, detection, and morphometry,” Nat. Methods 16, 67–70 (2019). https://doi.org/10.1038/s41592-018-0261-2
K. Han, Y. Wang, H. Chen, X. Chen, J. Guo, Z. Liu, Y. Tang, A. **ao, C. Xu, Y. Xu, Z. Yang, Y. Zhang, and D. Tao, “A survey on vision transformer” (2021). ar**v:2012.12556 [cs.CV]
S. Khan, M. Naseer, M. Hayat, S. W. Zamir, F. S. Khan, and M. Shah, “Transformers in vision: A survey,” ACM Comput. Surv. (2021). https://doi.org/10.1145/3505244
Y. Li, S. Wang, J. Wang, G. Zeng, W. Liu, Q. Zhang, Q. **, and Y. Wang, “GT U-Net: A U-Net like group transformer network for tooth root segmentation,” in Machine Learning in Medical Imaging. MLMI 2021, Ed. by C. Lian, X. Cao, I. Rekik, X. Xu, and P. Yan, Lecture Notes in Computer Science, vol. 12966 (Springer, Cham, 2021), pp. 386–395. https://doi.org/10.1007/978-3-030-87589-3_40
Z. Liu, Y. Lin, Y. Cao, H. Hu, Y. Wei, Z. Zhang, S. Lin, and B. Guo, “Swin transformer: Hierarchical vision transformer using shifted windows,” in Proc. IEEE/CVF Int. Conf. on Computer Vision (ICCV), 2021 (IEEE, 2021), pp. 10012–10022. ar**v:2103.14030 [Cs]
M. Niemeijer, J. Staal, B. van Ginneken, M. Loog, and M. D. Abramoff, “Comparative study of retinal vessel segmentation methods on a new publicly available database,” Proc. SPIE 5370, 648–656 (2004). https://doi.org/10.1117/12.535349
O. Ronneberger, P. Fischer, and T. Brox, U-Net: Convolutional Networks for Biomedical Image Segmentation, in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, Ed. by N. Navab, J. Hornegger, W. M. Wells, and A. F. Frangi, Lecture Notes in Computer Science, vol. 9351 (Springer, Cham, 2015), pp. 234–241. https://doi.org/10.1007/978-3-319-24574-4_28
A. Srinivas, T.-Y. Lin, N. Parmar, J. Shlens, P. Abbeel, and A. Vaswani, “Bottleneck transformers for visual recognition,” in IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, Tenn., 2021 (IEEE, 2021), pp. 16514–16524. https://doi.org/10.1109/CVPR46437.2021.01625
A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, Ł. Kaiser, and I. Polosukhin, “Attention is all you need,” in Advances in Neural Information Processing Systems, Ed. by I. Guyon, S. Vishwanathan, and R. Garnett (Curran Associates, 2017), pp. 5998–6008.
H. Wu, B. **ao, N. Codella, M. Liu, X. Dai, L. Yuan, and L. Zhang, “CvT: Introducing convolutions to vision transformers,” in Proc. IEEE/CVF Int. Conf. on Computer Vision (ICCV), 2021 (2021), pp. 22–31. ar**v:2103.15808 [cs]
The RIPS Dataset. https://www.icar.cnr.it/sites-rips-datasetrips/. Cited January 19, 2022.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
This article is a completely original work of its authors; it has not been published before and will not be sent to other publications until the PRIA Editorial Board decides not to accept it for publication.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information

Mara Sangiovanni received the master degree in Computer Science “cum laude” in 2009 and the Ph.D. degree in Bioinformatics and Computational Biology in 2014 at the University of Naples Federico II. She is a researcher at the National Research Council of Italy. Her research interests include computational biology and biomedical image processing. She has experience in machine learning applied to -omics data.

Nadia Brancati received her M.Sc. degrees in Computer Science “cum laude” at the University of Naples Parthenope in 2008. She is a researcher at the National Research Council of Italy. Her research interests include mainly image processing, computer vision and human computer interaction, in particular segmentation, image analysis, classification and medical imaging. She has experience in machine learning, with particular reference to deep learning. She is actively involved in international/national projects, even with industrial collaborations. She has participated in two international competitions (BACH 2018 and SSRBC 2017), resulting for both at the second position in the ranking. She is a member of the Italian Association for Computer Vision, Pattern Recognition and Machine Learning (CVPL).

Maria Frucci is senior researcher at the National Research Council (CNR) of Italy. She is head of the Artificial Intelligence in Image and Signal Analysis group at the Institute for High-Performance Computing and Networking (ICAR-CNR) carrying out design and development of Artificial Intelligence models and methods for the automatic analysis of images and signals in Medical/Biological Imaging and Healthcare. Maria has been Adjunct Professor in Computer Science at the University “Federico II” of Naples, teaching algorithms and data structures. She is a member of the Italian Association for Computer Vision, Pattern Recognition and Machine Learning (CVPL) and was member of the Governing Board of the Italian Group of Researchers in Pattern Recognition (CVPL-ex GIRPR). Before joining CNR, Maria worked as a researcher at the Centre for Informatics and Industrial Automation Research (CRIAI) where she was in charge of the Artificial Intelligence and Image Analysis research group of CRIAI that conducted numerous research activities in the field of Natural Language, Expert Systems and Shape Analysis. She received the doctoral degree “cum laude” in Physics from the University “Federico II” of Naples, Italy. Maria has been involved in the organization of several international conferences and has published more than 110 papers on different topics such as natural language, perception, computer vision, pattern recognition and deep learning. She has participated to two international competitions (BACH 2018 and SSRBC 2017), resulting for both at the second position in the ranking.

Luigi Di Perna received the Laurea degree in medicine and surgery from the Second University of Naples, Italy, in 2010, and the Residency in Ophthalmology cum laude in 2016. From 2014 to 2015, he held an Observership at Cliniques Universitaires Saint Luc, Bruxelles, Belgium. Mr. Di Perna is a current member of the Association for Research in Vision and Ophthalmology (ARVO). He received the travel grant at the 2016 ARVO Annual Meeting for the paper Activation of Melanocortin Receptors Mc1 and Mc5 Attenuates Retinal Damages in Experimental Diabetic Retinopathy.

Francesca Simonelli was born in Nola, Italy, in 1959. She received the degree (Hons.) in medicine and surgery from the University of Naples Federico II, Naples, Italy, in 1983, and the Residency in Ophthalmology from the School of Medicine, University of Naples Federico II, in 1987. She is currently a Full Professor of Ophthalmology with the Multidisciplinary Department of Medical, Surgical and Dental Sciences, University of Campania Luigi Vanvitelli, Naples. She has authored or coauthored about 100 journal and conference papers in the field of ophthalmology. Prof. Simonelli is a member of the Association for Research in Vision and Ophthalmology, the President of the Italian Society of Ophthalmologic Genetics, a Member of the Eye Working Group of the Telethon Institute of Genetics and Medicine, a Member of the National Fighting Blindness Committee of the Italian Ministry of Health, a Member of the Italian Society of Ophthalmology, and the President of the Scientific Committee of Retina Italia Onlus.

Daniel Riccio received the Laurea degree “cum laude” and the Ph.D. degree in Computer Sciences from the University of Salerno, Italy, in 2002 and 2006, respectively. He is currently an Associate Professor at the University of Naples Federico II. He is co-chief of the PRISCALab and his research interests include biometrics, medical imaging, image processing and indexing, image and video analytics. Daniel Riccio is member of the Artificial Intelligence, Privacy & Applications (AIPA) Lab working on machine learning, data science, biometrics and security applications. He is also an Associate Researcher at the National Research Council of Italy. Daniel Riccio is an IEEE member and a member of the Italian Association for Computer Vision, Pattern Recognition and Machine Learning (CVPL).
Rights and permissions
About this article
Cite this article
Sangiovanni, M., Brancati, N., Frucci, M. et al. Segmentation of Pigment Signs in Fundus Images with a Hybrid Approach: A Case Study. Pattern Recognit. Image Anal. 32, 312–321 (2022). https://doi.org/10.1134/S1054661822020171
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1054661822020171