Abstract
Patients with coronavirus disease 2019 (COVID-19) present increased risk for ischemic cardiovascular complications up to 1 year after infection. Although the systemic inflammatory response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection likely contributes to this increased cardiovascular risk, whether SARS-CoV-2 directly infects the coronary vasculature and attendant atherosclerotic plaques remains unknown. Here we report that SARS-CoV-2 viral RNA is detectable and replicates in coronary lesions taken at autopsy from severe COVID-19 cases. SARS-CoV-2 targeted plaque macrophages and exhibited a stronger tropism for arterial lesions than adjacent perivascular fat, correlating with macrophage infiltration levels. SARS-CoV-2 entry was increased in cholesterol-loaded primary macrophages and dependent, in part, on neuropilin-1. SARS-CoV-2 induced a robust inflammatory response in cultured macrophages and human atherosclerotic vascular explants with secretion of cytokines known to trigger cardiovascular events. Our data establish that SARS-CoV-2 infects coronary vessels, inducing plaque inflammation that could trigger acute cardiovascular complications and increase the long-term cardiovascular risk.
Similar content being viewed by others
Main
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is uniquely marked by extraordinary tissue tropism and an array of clinical presentations, from asymptomatic infection to acute respiratory distress, multi-organ failure and death1. Ischemic cardiovascular events, such as acute myocardial infarction (AMI) and stroke, due to the underlying disruption of a chronically inflamed atherosclerotic plaque2, are established clinical complications of COVID-19 (refs. 1,3). AMI and stroke can be triggered by several acute respiratory viral infections, including influenza virus4. However, patients with COVID-19 are >7-fold more likely to have a stroke than patients with influenza5, and their risk for both AMI and stroke remains high for up to 1 year after infection16. This analysis revealed that VSMCs in atherosclerotic lesions did not express significant levels of Cd68 and maintained a high level of Acta2 expression, although its expression was reduced compared to non-atherosclerotic conditions (Extended Data Fig. 2b). Based on these results, we used ACTA2 probe to identify VSMCs and macrophages of VSMC origin as ACTA2+, and we included probes for the S and S antisense vRNA in the analysis to identify ACTA2+ infected cells. Notably, this analysis identified S+ACTA2+ cells and S antisense+ACTA2+ cells in human coronaries (Extended Data Fig. 2c); however, the median of the frequency of SARS-CoV-2-infected VSMCs (~0.14%) in human coronary was ~8 times lower than that of infected macrophages (~1.2%). These results show that, although SARS-CoV-2 can infect VSMCs, the number of infected VSMCs in human coronaries was lower compared to the number of infected macrophages. To further investigate SARS-CoV-2 infection of VSMCs and lipid-laden VSMCs, which are associated with atherosclerosis17,18,19,20, we infected primary human aortic VSMCs, as well as VSMCs loaded with cyclodextrin–cholesterol complexes (Extended Data Fig. 2d), with the SARS-CoV-2 USA WA1/2020 isolate. Approximately 18% of cultured VSMCs and 13% of cholesterol-loaded VSMCs were S+, and the frequency of S antisense+ACTA2+ cells, indicating viral replication, was ~2.6% (Extended Data Fig. 2d,e). Taken together with our in vitro findings, which indicate that more than 79% of macrophages and over 90% of foam cells are S+, along with the discovery that more than 40% of both cell types are S antisense+, these results show that, although SARS-CoV-2 can infect VSMCs, macrophages are infected at a higher rate.
SARS-CoV-2 infection of human macrophages and foam cells
The accumulation of cholesterol-laden macrophages (foam cells) is a hallmark of atherosclerosis at all stages of the disease, from early PIT to late fibroatheroma lesions10,12. To investigate SARS-CoV-2 infection of both macrophages and foam cells, we differentiated human monocytes derived from human peripheral blood mononuclear cells into macrophages and treated them with oxidized low-density lipoprotein (oxLDL) complexed with Dil dye (Dil-Ox-LDL) to differentiate them into foam cells. To experimentally confirm our observation that SARS-CoV-2 can infect human plaque macrophages, macrophages and foam cells were infected either with icSARS-CoV-2 mNeonGreen (mNG) reporter virus, a modified virus that allows the use of mNG fluorescence as a surrogate readout for viral replication16 were extracted from BioProject accession number PRJNA626450. Quality control of scRNA-seq data was performed using FastQC (version 0.11.7). Reads were aligned to the GRCm39 (mm39) reference genome using STAR (version 2.6.1d). FeatureCounts from the subread package (version 1.6.3) was employed and normalized counts used for downstream analysis using the Seurat R package (version 4.3.0). Highly variable genes were identified using the FindVariableFeatures function. RunUMAP function with default settings was used with FindNeighbors and FindClusters functions for cell clustering. Differential gene expression analysis was performed using the FindMarkers function to identify differences between single-color Tomato reporter (Myh11-CreERT2, Rosa26tdTomato/tdTomato, ApoE−/− mice) fed a high-fat diet (21% anhydrous milk fat, 19% casein and 0.25% cholesterol) for 18 weeks versus control mice. The Benjamini–Hochberg method was applied to control for the false discovery rate (FDR).
Experiments in Biosafety Level 3
Studies involving SARS-CoV-2 infection were approved by the Institutional Biosafety Committee (IBC21-000079) of the NYU Grossman School of Medicine. All Biosafety Level 3 procedures were conducted in accordance with the Biosafety Manual and standard operating procedures of the NYU Grossman School of Medicine High-Containment Facility.
Cells and viruses
Vero E6 cells (American Type Culture Collection, CRL-1586) were maintained in DMEM culture media containing 10% FBS (Gibco), 2 mM l-glutamine and 100 U ml−1 penicillin–streptomycin. Vero E6 Expressing Transmembrane Protease, Serine 2 and Human Angiotensin-Converting Enzyme 2 (Vero E6-TMPRSS2-T2A-ACE2) were obtained from BEI Resources (NR-54970). Vero E6-TMPRSS2-T2A-ACE2 cells were grown in DMEM medium with 4 mM l-glutamine, 4,500 ml of glucose, 1 mM sodium pyruvate and 1,500 mg L−1 sodium bicarbonate, 10% FBS and 10 μg ml−1 puromycin. All cells were verified to be free of mycoplasma contamination.
SARS-CoV-2 isolate USA-WA1/2020 (BEI Resources, NR52281) was amplified once in Vero E6 cells infected at a multiplicity of infection (MOI) of 0.01 as previously described69. Virus was collected at 72 hpi upon observation of cytopathic effect. Debris was removed by centrifugation and passage through a 0.22-μm filter, and the supernatant was then aliquoted and stored at −80 °C. Virus titer was calculated by plaque assay on Vero E6 cells and informed as particle-forming units per milliliter (PFU ml−1). Virus stocks were Sanger sequenced during viral stock production. A PCR amplicon covering the S gene (FW: gttcagagtttattctagtgcgaataattgcacttttg, RV: gcagtaaggatggctagtgtaactagcaagaataccac) was purified using the Nucleospin PCR and Gel Extraction Kit (Macherey-Nagel) and Sanger sequenced (GENEWIZ) with the following primers (FW: ggttttaattgttactttcc and FW: ctacaggttctaatgtttttc). icSARS-CoV-2 mNG reporter virus was obtained from the UTMB World Reference Center for Emerging Viruses and ArbovirusesBulk RNA-seq RNA from primary macrophages and foam cells was extracted using TRIzol reagent and Direct-zol RNA Microprep Kits following the manufacturer’s instructions. Total RNA from human atherosclerotic tissue was isolated using QIAzol Lysis Reagent (Qiagen) and the gentleMACS Octo Dissociator (Miltenyi Biotec) homogenizer, combined with RNA cleanup using the RNAeasy Mini Kit (Qiagen). Quality control was performed with Agilent RNA 6000 Nano and Pico Kits (Agilent Technologies) using the Agilent 2100 Bioanalyzer system. For in vitro experiments, poly(A) library preparation was performed using Illumina Stranded mRNA Preparation and Ligation (Illumina). For human atherosclerotic plaque ex vivo experiments, the Revelo RNA-Seq High Sensitivity library preparation kit was used (Tecan). Libraries were quantified using KAPA Library Quantification Kit (Roche), pooled at 2 nM equimolar concentration and sequenced using an Illumina NovaSeq 6000 sequencer. Quality control of RNA-seq data was performed using FastQC2 (version 0.11.7). Raw sequenced reads were trimmed using fastp3 (version 0.20.1) for quality control of bases and to eliminate sequencing adaptors. Raw reads were aligned using STAR (version 2.6.1d) to the combined human (Homo sapiens) genome assembly GRCh38 from the Genome Reference Consortium (GCA_000001405.15 GCF_000001405.26) and SARS-CoV-2 Washington isolate (USA WA1/2020) genome (GenBank: MN985325.1). The gene-level expression counts were computed with the featureCounts function in the Subread package (version 1.6.3; parameters: -g gene_id -s 2) using the human gene annotations from GENCODE release 33. Differential expression was performed using the R package DESeq2 (version 1.30.1). To model differences in gene expression between SARS-CoV-2-infected versus non-infected primary macrophages and foam cells, a model including infection status, timepoint and donor as dependent variables was used. To identify differences in gene expression between infected macrophages and infected foam cells, a model including cell type, timepoint and donor as dependent variables was used. To analyze gene expression variation across timepoints and infection status in macrophages and foam cells, we employed a model that incorporated infection status, timepoint, donor and an interaction between infection status and timepoint as dependent variables for each sample type separately. The IFN and SARS-CoV-2 scores were calculated as log2 values of IFN response genes and SARS-CoV-2 genes comparing macrophage and foam cell response at 0 hpi, 2 hpi, 8 hpi, 24 hpi and 48 hpi. Standardized data (z-scores) were calculated for each feature by subtracting the estimate mean and dividing by the estimate s.d. For hierarchical clustering, data were plotted using the pheatmap package (version 1.0.12) in R. Differential expression analysis of SARS-CoV-2-infected atherosclerotic plaque samples was performed using the R package DESeq2 with timepoint and donor included as dependent variables. For hierarchical clustering analysis, normalized values were standardized and plotted using the pheatmap package (version 1.0.12) in R. P values were adjusted using Benjamini–Hochberg correction and denoted as an asterisk. Gene set enrichment analysis using Reactome Knowledgebase 2022 and Gene Ontology Biological Process 2021 of the top 300 DEGs was performed using Enrichr (https://maayanlab.cloud/Enrichr/)70,71. Bar plots represent the combined score of 10 top relevant pathways with statistical significance (*P < 0.05, **P < 0.01; ***P < 0.001). A screen of 48 human cytokines and chemokines was performed using UV-inactivated culture supernatants by using the Bio-Plex Pro Human Cytokine Screening Panel (Bio-Rad) and the Luminex 200 platform of the Immune Monitoring Laboratory Division of Advanced Research Technologies of the NYU Grossman School of Medicine. Luminex data were log transformed; statistically significant differences were calculated using unpaired two-sided t-tests; and P values were adjusted using Benjamini–Hochberg correction. Empirical Bayes batch correction (Combat) was used to remove batch effects before log transforming the data. Statistical analysis was performed using R (version 4.0.3). Cytokines showing log2FC > 0 were upregulated, and cytokines showing log2FC < 0 were downregulated. Secreted TGF-β1 and Caspase-8 were measured by ELISA (Invitrogen) in clarified culture media supernatant, according to the manufacturer’s instructions. After 48 hpi and 72 hpi, ex vivo SARS-CoV-2-infected atherosclerotic samples were fixed with 3% glutaraldehyde/PBS (pH 7.4) at 4 °C. Samples were prepared for electron microscopic evaluation by the NYU Grossman School of Medicine’s Microscopy Laboratory following standard operating procedures. The specimens were examined by transmission electron microscopy. Stained grids were imaged with a Talos L120C transmission electron microscope and recorded using a Gatan OneView Camera (4 K × 4 K resolution) with Digital Micrograph software (Gatan Microscopy Suite). Statistical analyses not described above were performed using GraphPad Prism version 9.0, and details are included in the figure legends. Statistical P values were calculated and reported on graphs, and P < 0.05 was considered significant. Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.RNA-seq data processing, analysis and visualization
Cytokine and chemokine protein secretion
Transmission electron microscopy
Statistical analysis
Reporting summary
Data availability
scRNA-seq data from the six human carotid arteries were previously published and are available in the GEO (GSE224273). Four additional carotid plaques were processed to obtain sc-RNAseq data deposited in the GEO (GSE235437). scRNA-seq data coronary lesions were obtained from the GEO (GSE131780). Bulk RNA-seq data from macrophages and foam cell experiments and the plaque tissues infected with SARS-CoV-2 are deposited in the GEO (GSE235437). GTEx data used are version V8, dbGaP accession number phs000424.v8.p2, and available at the GTEx portal (https://gtexportal.org/home). Murine scRNA-seq data from ref. 16 were extracted from BioProject accession number PRJNA626450. Human (Homo sapiens) genome assembly GRCh38 (hg38) was obtained from the Genome Reference Consortium (GCA_000001405.15 GCF_000001405.26). SARS-CoV-2 Washington isolate (WA1/2020) genome was obtained from the NIH repository (GenBank: MN985325.1). The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/USA-WA1/2020, NR-52281. The following reagent was obtained through BEI Resources, NIAID, NIH Cercopithecus aethiops Kidney Epithelial Cells Expressing Transmembrane Protease, Serine and Human Angiotensin-Converting Enzyme 2 (Vero E6-TMPRSS2-T2A-ACE2, NR-54970). All other data supporting the findings in this study are included in the main article and associated files. Source data files are available on GitHub at https://github.com/giannarelli-lab/SARS-CoV-2-infection-triggers-pro-atherogenic-inflammatory-responses-in-human-coronary-vessels.
Code availability
Code used for data analysis is available on GitHub at https://github.com/giannarelli-lab/SARS-CoV-2-infection-triggers-pro-atherogenic-inflammatory-responses-in-human-coronary-vessels.
References
Lamers, M. M. & Haagmans, B. L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 20, 270–284 (2022).
Engelen, S. E., Robinson, A. J. B., Zurke, Y. X. & Monaco, C. Therapeutic strategies targeting inflammation and immunity in atherosclerosis: how to proceed? Nat. Rev. Cardiol. 19, 522–542 (2022).
Katsoularis, I., Fonseca-Rodriguez, O., Farrington, P., Lindmark, K. & Fors Connolly, A. M. Risk of acute myocardial infarction and ischaemic stroke following COVID-19 in Sweden: a self-controlled case series and matched cohort study. Lancet 398, 599–607 (2021).
Kwong, J. C. et al. Acute myocardial infarction after laboratory-confirmed influenza infection. N. Engl. J. Med. 378, 345–353 (2018).
Merkler, A. E. et al. Risk of ischemic stroke in patients with Coronavirus Disease 2019 (COVID-19) vs patients with influenza. JAMA Neurol. 77, 1366–1372 (2020).
**e, Y., Xu, E., Bowe, B. & Al-Aly, Z. Long-term cardiovascular outcomes of COVID-19. Nat. Med. 28, 583–590 (2022).
Fajgenbaum, D. C. & June, C. H. Cytokine storm. N. Engl. J. Med. 383, 2255–2273 (2020).
Liu, J. et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 7, 17 (2021).
Sefik, E. et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature 606, 585–593 (2022).
Moore, K. J., Sheedy, F. J. & Fisher, E. A. Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol. 13, 709–721 (2013).
Narula, N., Olin, J. W. & Narula, N. Pathologic disparities between peripheral artery disease and coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 40, 1982–1989 (2020).
Otsuka, F. et al. Natural progression of atherosclerosis from pathologic intimal thickening to late fibroatheroma in human coronary arteries: a pathology study. Atherosclerosis 241, 772–782 (2015).
Saccon, T. D. et al. SARS-CoV-2 infects adipose tissue in a fat depot- and viral lineage-dependent manner. Nat. Commun. 13, 5722 (2022).
Martinez-Colon, G. J. et al. SARS-CoV-2 infection drives an inflammatory response in human adipose tissue through infection of adipocytes and macrophages. Sci. Transl. Med. 14, eabm9151 (2022).
Wirka, R. C. et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat. Med. 25, 1280–1289 (2019).
Wang, Y. et al. Clonally expanding smooth muscle cells promote atherosclerosis by esca** efferocytosis and activating the complement cascade. Proc. Natl Acad. Sci. USA 117, 15818–15826 (2020).
Rong, J. X., Shapiro, M., Trogan, E. & Fisher, E. A. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc. Natl Acad. Sci. USA 100, 13531–13536 (2003).
Shankman, L. S. et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat. Med. 21, 628–637 (2015).
Feil, S. et al. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ. Res. 115, 662–667 (2014).
Allahverdian, S., Chehroudi, A. C., McManus, B. M., Abraham, T. & Francis, G. A. Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation 129, 1551–1559 (2014).
**e, X. et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27, 841–848 (2020).
Combe, M., Garijo, R., Geller, R., Cuevas, J. M. & Sanjuan, R. Single-cell analysis of RNA virus infection identifies multiple genetically diverse viral genomes within single infectious units. Cell Host Microbe 18, 424–432 (2015).
Heldt, F. S., Kupke, S. Y., Dorl, S., Reichl, U. & Frensing, T. Single-cell analysis and stochastic modelling unveil large cell-to-cell variability in influenza A virus infection. Nat. Commun. 6, 8938 (2015).
Zhu, Y., Yongky, A. & Yin, J. Growth of an RNA virus in single cells reveals a broad fitness distribution. Virology 385, 39–46 (2009).
Cohen, E. M., Avital, N., Shamay, M. & Kobiler, O. Abortive herpes simplex virus infection of nonneuronal cells results in quiescent viral genomes that can reactivate. Proc. Natl Acad. Sci. USA 117, 635–640 (2020).
Ioannidis, L. J., Verity, E. E., Crawford, S., Rockman, S. P. & Brown, L. E. Abortive replication of influenza virus in mouse dendritic cells. J. Virol. 86, 5922–5925 (2012).
Lucas, C. et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–469 (2020).
Munnur, D. et al. Altered ISGylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat. Immunol. 22, 1416–1427 (2021).
Cheng, J. et al. IL-27 induces IFN/STAT1-dependent genes and enhances function of TIGIT+ HIVGag-specific T cells. iScience 25, 103588 (2022).
Heaton, N. S. & Randall, G. Multifaceted roles for lipids in viral infection. Trends Microbiol 19, 368–375 (2011).
Cloherty, A. P. M., Olmstead, A. D., Ribeiro, C. M. S. & Jean, F. Hijacking of lipid droplets by hepatitis C, dengue and Zika viruses—from viral protein moonlighting to extracellular release. Int. J. Mol. Sci. 21, 7901 (2020).
Theken, K. N., Tang, S. Y., Sengupta, S. & FitzGerald, G. A. The roles of lipids in SARS-CoV-2 viral replication and the host immune response. J. Lipid Res. 62, 100129 (2021).
Collins, K. L., Chen, B. K., Kalams, S. A., Walker, B. D. & Baltimore, D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391, 397–401 (1998).
Hansen, T. H. & Bouvier, M. MHC class I antigen presentation: learning from viral evasion strategies. Nat. Rev. Immunol. 9, 503–513 (2009).
Bizzotto, J. et al. SARS-CoV-2 infection boosts MX1 antiviral effector in COVID-19 patients. iScience 23, 101585 (2020).
Domizio, J. D. et al. The cGAS–STING pathway drives type I IFN immunopathology in COVID-19. Nature 603, 145–151 (2022).
Prelli Bozzo, C. et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 12, 4584 (2021).
Kuznetsova, T., Prange, K. H. M., Glass, C. K. & de Winther, M. P. J. Transcriptional and epigenetic regulation of macrophages in atherosclerosis. Nat. Rev. Cardiol. 17, 216–228 (2020).
Seth, R. B., Sun, L., Ea, C. K. & Chen, Z. J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122, 669–682 (2005).
Minkoff, J. M. & tenOever, B. Innate immune evasion strategies of SARS-CoV-2. Nat. Rev. Microbiol. 21, 178–194 (2023).
Garcia-Nicolas, O., Godel, A., Zimmer, G. & Summerfield, A. Macrophage phagocytosis of SARS-CoV-2-infected cells mediates potent plasmacytoid dendritic cell activation. Cell. Mol. Immunol. 20, 835–849 (2023).
Janeway, C. A.Jr & Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 (2002).
tenOever, B. R. The evolution of antiviral defense systems. Cell Host Microbe 19, 142–149 (2016).
Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045 (2020).
Lazear, H. M., Schoggins, J. W. & Diamond, M. S. Shared and distinct functions of type I and type III interferons. Immunity 50, 907–923 (2019).
Ridker, P. M. & Rane, M. Interleukin-6 signaling and anti-interleukin-6 therapeutics in cardiovascular disease. Circ. Res. 128, 1728–1746 (2021).
Ridker, P. M. et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 377, 1119–1131 (2017).
Zernecke, A., Bernhagen, J. & Weber, C. Macrophage migration inhibitory factor in cardiovascular disease. Circulation 117, 1594–1602 (2008).
Wang, J. et al. Interleukin 18 function in atherosclerosis is mediated by the interleukin 18 receptor and the Na-Cl co-transporter. Nat. Med. 21, 820–826 (2015).
Bastard, P. et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, eabd4585 (2020).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280 (2020).
Kyrou, I., Randeva, H. S., Spandidos, D. A. & Karteris, E. Not only ACE2—the quest for additional host cell mediators of SARS-CoV-2 infection: neuropilin-1 (NRP1) as a novel SARS-CoV-2 host cell entry mediator implicated in COVID-19. Signal Transduct. Target. Ther. 6, 21 (2021).
Cantuti-Castelvetri, L. et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860 (2020).
Daly, J. L. et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370, 861–865 (2020).
Dann, E., Henderson, N. C., Teichmann, S. A., Morgan, M. D. & Marioni, J. C. Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol. 40, 245–253 (2022).
Kim, K. et al. Transcriptome analysis reveals nonfoamy rather than foamy plaque macrophages are proinflammatory in atherosclerotic murine models. Circ. Res. 123, 1127–1142 (2018).
Solomon, B. D., Mueller, C., Chae, W. J., Alabanza, L. M. & Bynoe, M. S. Neuropilin-1 attenuates autoreactivity in experimental autoimmune encephalomyelitis. Proc. Natl Acad. Sci. USA 108, 2040–2045 (2011).
Dai, X. et al. A novel role for myeloid cell-specific neuropilin 1 in mitigating sepsis. FASEB J. 31, 2881–2892 (2017).
Stein, S. R. et al. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature 612, 758–763 (2022).
Lindner, D. et al. Association of cardiac infection with SARS-CoV-2 in confirmed COVID-19 autopsy cases. JAMA Cardiol. 5, 1281–1285 (2020).
Robbins, C. S. et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat. Med. 19, 1166–1172 (2013).
Bajpai, G. et al. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat. Med. 24, 1234–1245 (2018).
Brewer, C. M. & Majesky, M. W. Branch point smooth muscle cells highlighted by novel lineage tracking approach. Circ. Res. 122, 194–196 (2018).
Nilsson-Payant, B. E. et al. The NF-κB transcriptional footprint is essential for SARS-CoV-2 replication. J. Virol. 95, e0125721 (2021).
Wang, S. et al. Programmed death ligand 1 expression and tumor infiltrating lymphocytes in neurofibromatosis type 1 and 2 associated tumors. J. Neurooncol. 138, 183–190 (2018).
Fernandez, D. M. et al. Single-cell immune landscape of human atherosclerotic plaques. Nat. Med. 25, 1576–1588 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Noval, M. G. et al. Antibody isotype diversity against SARS-CoV-2 is associated with differential serum neutralization capacities. Sci Rep. 11, 5538 (2021).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14, 128 (2013).
Acknowledgements
We thank the NYU Biosafety Level 3 High-Containment Facility, the NYULH Center for Biospecimen Research and Development, the Histology and Immunohistochemistry Laboratory (CBRD; RRID:SCR_018304), the Experimental Pathology Division (ExPath), the NYU Genome Technology Center (GTC), the NYU Microscopy Laboratory and the NYU Immune Monitoring Laboratory (IML) at NYU Langone’s Division of Advanced Research Technologies (DART) for their assistance. The DART CBRD, ExPath, IML and the Microscopy Lab cores are supported by NIH/NCI grant P30CA016087. CBRD is also supported by the Laura and Isaac Perlmutter Cancer Center Support Grant. ExPath is also supported by NIH S10 OD021747. This work was funded by NIH/NHLBI grant 1R01HL165258 (C.G.). C.G. also acknowledges support from grants NIH/NHLBI R01HL153712, AHA 20SFRN35210252, CZI NFL-2020-218415 and U34TR003594. N.E is supported by the AHA research supplement to promote diversity in science (AHA 965509). M.G.N is supported by AHA Postdoctoral Fellowship 19-A0-00-1003686. D.D. is supported by AHA grant 20SFRN35210252. SARS-CoV-2 work in the M.S. laboratory is supported by NIH/NIAID R01AI160706 and NIH/NIDDK R01DK130425. K.A.S. is supported by NIH/NIAID 1R01 AI162774-01A1, NYU Grossman School of Medicine startup funds and an NYU Cardiovascular Research Center pilot award. K.J.M. is supported by NIH/NHLBI R35HL, R35HL135799 and R01HL084312. The publicly available data used for the analyses described in this paper were obtained from the GTEx portal. The GTEx project was supported by the Common Fund of the Office of the Director of the National Institutes of Health and by NCI, NHGRI, NHLBI, NIDA, NIMH and NINDS.
Author information
Authors and Affiliations
Contributions
Conceptualization: N.E. and C.G. Methodology: N.E., M.G.N., O.S., R.K., K.M, K.A.S., N.N. and L.A. Biosafety Level 3 experiments: N.E., M.G.N. and S.J. Other experiments: M.S., N.E., O.S, B.C., L.A., D.D. and A.V.G. Patient recruitment: R.S., S.S. and D.F. Clinical data management: N.E., S.S., J.N., N.N., R.S., P.F. and A.R. Human sample collection: P.F., N.N., A.R., T.M. and C.R. Human sample processing: N.E., L.A., A.R., D.F., B.C. and D.D. Data analysis: N.E., D.D., M.G.N., R.K., M.G., D.F., J.N., B.C., D.D. and L.A. Resources: C.G., K.A.S. and A.R. Data visualization: N.E., R.K., M.G. and L.A. Writing—original draft: N.E. and C.G. Writing—revision and editing: all authors. Supervision: C.G. Project administration and funding acquisition: C.G.
Corresponding author
Ethics declarations
Competing interests
The M.S. laboratory has received unrelated research funding in sponsored research agreements from ArgenX N.V., Moderna and Phio Pharmaceuticals, which has no competing interest with this work. The authors declare no other competing interests.
Peer review
Peer review information
Nature Cardiovascular Research thanks Peter Libby, Claudia Monaco and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Elvira Forte, in collaboration with the Nature Cardiovascular Research team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended Data
Extended Data Fig. 1 Pathology assessment and RNA-FISH analysis of coronary arteries from deceased individuals with COVID-19.
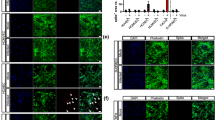
a) Bar plot (left) shows the number of adaptive intimal thickening (AIT; blue, n = 4), pathological intimal thickening (PIT; magenta, n = 10), fibrocalcific (orange, n = 10) and fibroatheroma (red, n = 3) specimens. Bar plot (right) shows the proportion of coronaries that presented pathological features of lipid pool, calcification, necrotic core, adventitial inflammation, and thrombus. b) Bar plot showing the percentage of CD68+ area. Non-parametric Kruskal-Wallis test with Dunn’s test for multiple comparisons was performed. c) Bar plot showing the quantification of frequency and total count of CD68+ cells in AIT (n = 4), PIT (n = 10), fibrocalcific (n = 8) and fibroatheroma (n = 3). Data are presented as mean values ± s.e.m. Non-parametric Kruskal-Wallis test with Dunn’s test for multiple comparisons was performed. d) Bar plots of total cell number normalized by the area (mm2) of vasculature and perivascular fat. Each dot represents a tissue section from AIT (n = 12), PIT (n = 15), fibrocalcific (n = 24) and fibroatheroma (n = 9). Data are presented as mean values ± s.e.m. One-way ANOVA with post-hoc Tukey’s test for multiple comparisons was performed. e) Bar plots of total number of CD68 RNA+ cells quantified in the arterial wall and perivascular fat. Dots represent individual tissue sections from AIT (n = 6), PIT (n = 12), fibrocalcific (n = 16) and fibroatheroma (n = 6). Data are presented as mean values ± s.e.m. One-way ANOVA followed by Holm-Šídák’s test for multiple comparisons was performed. f) Bar plots of frequency of CD68+ SARS-CoV-2 Spike+ and CD68+ Spike antisense+ cells in AIT (n = 3), PIT (n = 6), fibrocalcific (n = 8) and fibroatheroma (n = 3) coronaries. Non-parametric Kruskal-Wallis test followed by uncorrected Dunn’s test for multiple comparisons was performed. g) Scatter plot of Spearman’s rank correlation (95% confidence interval) of total CD68 RNA copies with total SARS-CoV-2 spike and Spike antisense copies (n = 20). h) SARS-CoV-2 S and S antisense quantification in total tissue, vasculature, and perivascular fat from patients with (n = 7 samples) versus without CV manifestations (n = 13 samples). Data were normalized by tissue area (mm2) and presented as mean values ± s.e.m. Rout method (Q = 1%) was used to remove outliers. Unpaired Mann-Whitney test was performed.
Extended Data Fig. 2 Analysis of SARS-CoV-2 tropism for human vascular smooth muscle cells.
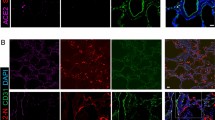
a) Differential gene expression analysis of ACTA2 and CD68 in vascular smooth muscle cells (VSMCs) and myeloid cell clusters from seven atherosclerotic coronary samples. Wilcoxon Rank-Sum test was used to compare groups and adjusted p-values with Benjamini-Hochberg correction method are shown. b) Differential gene expression analysis of Acta2 mRNA and Cd68 mRNA in lineage-tagged (Tdt+) SMC-derived cells dissociated from the aortic arches of atherosclerotic single-color Tomato reporter (Myh11-CreERT2, Rosa26tdTomato/tdTomato, ApoE−/− mice) mice fed high fat diet (HFD) for 18 weeks versus control mice. Wilcoxon Rank-Sum test was used to compare groups and adjusted p-values with BH correction method are shown. c) Representative images of spatial analysis of PIT coronary samples, and fibrocalcific and fibroatheroma (FCFA) samples showing the location of ACTA2+ cells, ACTA2+ SARS-CoV-2 Spike+ cells or ACTA2+ SARS-CoV-2 Spike antisense+ cells. Bar plots showing the number and frequencies of ACTA2+ cells/mm2, ACTA2+SARS-CoV-2 Spike vRNA+ and ACTA2+ SARS-CoV-2 S antisense+ cells normalized by tissue area (mm2) in intimal thickening (PIT; n = 6) versus fibrocalcific and fibroatheroma (n = 3) coronaries. Mann-Whitney test (two-tailed) was performed. d) Representative images of Oil Red-O staining of human VSMCs treated with 10 µg/mL of Cholesterol–methyl-β-cyclodextrin (Chol:MβCD) or vehicle overnight before infection and kept for 48 h. e) Bar plots showing the frequencies of SARS-CoV-2 Spike vRNA+, ACTA2+SARS-CoV-2 Spike vRNA+, SARS-CoV-2 S antisense+ and ACTA2+ SARS-CoV-2 Spike antisense+ cells in vehicle and Chol:MβCD conditions after infection with SARS-CoV-2 USA-WA1/2020 for 24 h. n = 20 of vehicle and n = 25 Chol:MβCD treated VSMCs in Spike vRNA quantification experiment, n = 21 of vehicle and n = 24 Chol:MβCD treated VSMCs in Spike antisense quantification experiment. Mann-Whitney test (two-tailed) was used.
Extended Data Fig. 3 SARS-CoV-2 in-vitro infection of human primary macrophages and foam cells.
a) Representative images of not-infected and infected macrophages and foam cells cultured with mNG reporter virus (MOI 0.1) at 24 hpi. Scale bar, 20 μm. b) Representative images of not-infected and SARS-CoV-2 infected macrophages and foam cells at 24 hours post-infection (hpi). Scale bar, 20 μm. c) Representative images of plaque assay in VERO E6-TMPRSS2-T2A-ACE2 cells of culture supernatants of macrophages and foam cells cultured with SARS-CoV-2 USA- WA1/2020 at 2 hpi, 8 hpi, 24 hpi, and 48 hpi. Serial dilutions are represented from left to right (-1 to -6). d) Bar plot showing the log10 FC of SARS-CoV-2 NP RNA levels normalized by 2 hpi samples (n = 4 biological replicates) in infected macrophages and foam cells. e) Bar plots showing the combined score of Gene Ontology (GO) Biological Process 2021 enrichment analysis of upregulated genes in infected macrophages, foam cells and or both vs. non-infected counterparts. P < 0.05. *, P < 0.05; **, P < 0.01; ***, P < 0.001. f) Heat map of log2 FC in complement genes between SARS-CoV-2 infected macrophages (n = 3) and SARS-CoV-2 infected foam cells (n = 3) at 0 hpi, 2 hpi, 8 hpi, 24 hpi and 48 hpi. Adjusted P-values < 0.05 (FDR = 10%) were considered significant. g) Heat map of Log2 fold changes in lysosomal genes in SARS-CoV-2 infected cells vs. not infected cells at 2 hpi, 8 hpi, 24 hpi and 48 hpi. h) Heat map of Log2 fold changes in lysosomal genes in SARS-CoV-2 infected macrophages vs. foam cells at 2 hpi, 8 hpi, 24 hpi and 48 hpi. P-values < 0.05 were considered significant. P < 0.05. *, P < 0.05; **, P < 0.01; ***, P < 0.001. i) Heat map of log2 FC in lipid metabolism genes in SARS-CoV-2 infected macrophages (n = 3) vs. foam cells (n = 3) at 2 hpi, 8 hpi, 24 hpi and 48 hpi. Adjusted P-values < 0.05 (FDR = 0.1) were considered significant. P < 0.05. *, P < 0.05; **, P < 0.01; ***, P < 0.001. j) Quantification of Caspase-8 concentration in culture supernatants of not infected or SARS-CoV-2 infected macrophages and foam cells. One-way ANOVA with post-hoc Tukey’s test were performed.
Extended Data Fig. 4 Dynamics of cytokine response in macrophages and foam cells after SARS-CoV-2 infection.
a) Heat maps of log2 FC of selected differentially expressed cytokine and chemokine genes in SARS-CoV-2 infected macrophages (left, n = 3) and foam cells (right, n = 3) versus. non-infected counterparts at different hpi. P-values were adjusted using Benjamini-Hochberg correction (FDR = 10%). Adjusted P-values < 0.05 were considered significant. Asterisk indicates an adjusted P-value < 0.05 for the comparison of infected vs not infected at each timepoint. Asterisk in parentheses indicates an adjusted P value < 0.05 for the interaction term of the model. *, P < 0.05; **, P < 0.01; ***, P < 0.001. b) Kinetic plots showing the area under the curve (AUC) of cytokines and chemokines in the supernatant of SARS-CoV-2 infected and non-infected macrophages and foam cells (n = 4 biological replicates, technical duplicates). For AUC comparisons, one-way ANOVA with Tukey’s test for multiple comparisons was used. Bar plots represent Mean ± s.e.m. One-way ANOVA followed by Šídák’s test for multiple comparisons was performed.
Extended Data Fig. 5 Ex-vivo SARS-CoV-2 infection of human carotid vascular explants.
a) Representative images of human atherosclerotic plaque tissues infected ex vivo with SARS-CoV-2 USA-WA1/2020 (105 PFU) versus mock infected control shows the expression of spike protein and nucleoprotein (NP). Scale bar, 100 μm. b) Electron microscopy of human atherosclerotic carotid plaque tissue infected ex vivo with the SARS-CoV-2. Scale bar, 1 μm. Black arrows indicate coronavirus-like particles. c) Heat map of selected cytokine and chemokine genes showing the log2 FC in SARS-CoV-2 infected carotid vascular explants versus not-infected tissues at different times post-infection. Wald test from DESeq2 package was used to test for significance. P values were adjusted using Benjamini-Hochberg correction (FDR = 10%) and denoted as an asterisk *, P < 0.05; **, P < 0.01; ***, P < 0.001. d) Kinetic plots showing the AUC of selected cytokines and chemokines secreted by non-infected or SARS-CoV-2 infected carotid vascular explants (n = 3 donors, technical duplicates) at different time post-infection. One-way ANOVA with Šídák’s test for multiple comparisons was performed. Bar plots represent mean ± s.e.m. Unpaired t-test (two-tailed) was performed and P < 0.05 was considered significant.
Extended Data Fig. 6 Single cell RNA sequencing analysis of SARS-CoV-2 entry factors in vascular myeloid subclusters.
a) Heat map shows transcripts expression (median TPM, transcripts per million) of SARS-CoV-2 entry factors identified in lung, whole blood, heart (left ventricle and atrial appendage), aorta, and tibial and coronary arteries. b) Violin plots showing the log10 TMP + 1 of tissue level expression of SARS-CoV-2 entry factors in lung, aorta, tibial and coronary artery identified. Data are presented as median ±IQR (25%-75% quartiles) in the box plot, violin plot defines density of data in whole range. c) UMAP embedding of integrated total immune cells from carotid (n = 10) and coronary (n = 7) tissues. d) Gene expression of SARS-CoV-2 viral entry factors and related genes projected onto the UMAP of total immune cells. e) UMAP representation of myeloid cell clusters colored by tissue origin. Dots represent individual cells belonging to carotid (red) or coronary artery (blue). f) Heatmap displaying selected z-score scaled genes (columns) across myeloid cell subclusters (rows) from human coronary and carotid samples. Canonical genes were used for myeloid subclusters annotations.
Extended Data Fig. 7 Abrogation of SARS-CoV-2 interaction with host by NRP-1 small molecule inhibitor and silencing RNA.
a) Dot plot showing the relative expression levels of NRP1 RNA normalized by GAPDH RNA expression in macrophages and foam cells. Average percentage of NRP1 silencing efficacy were calculated and depicted at the top (n = 4 biological replicates measured by technical duplicate per cell type, condition). b) Representative image of capillary western blot (Wes) was performed to evaluate the protein expression levels of NRP1 after siRNA NRP1 or siRNA control treatment. Target protein NRP1 (130-140 kD) and β-actin loading control blots (42 kD) are shown. c) Total NRP1 RNA copies were quantified in not-infected macrophages and foam cells treated with either siRNA control or siRNA NRP1 (n = 31 images of macrophages siRNA control; n = 26 macrophages siRNA NRP1, n = 24 foam cells siRNA control, n = 25 foam cells siRNA NRP1) at 24 hpi. d) Representative images and quantification of RNA-FISH showing NRP1 RNA in not-infected macrophages and foam cells. e) Representative images of RNA-FISH showing SARS-CoV-2 spike vRNA and NRP1 RNA (left), SARS-CoV-2 spike antisense vRNA and NRP1 RNA (right) in infected macrophages and foam cells treated with non-targeting siRNA control or siRNA NRP1 at 24 hpi. f) Representative images of RNA-FISH showing SARS-CoV-2 S vRNA and NRP1 RNA (left), SARS-CoV-2 S antisense RNA and NRP1 RNA (right) in infected macrophages and foam cells with and without NRP1-blocking (EG00229 trifluoroacetate) at 24 hpi.
Extended Data Fig. 8 Dynamics of cytokine response in NRP1- blocking/silencing treated macrophages and foam cells.
a) Heat map of standardized z-scored gene expression of cytokines and chemokines in not-infected, SARS-CoV-2 infected macrophages and foam cells treated with non-targeting siRNA control or siRNA NRP1 at 24 hpi. b) Quantification of TGF-β1 concentration (pg mL−1) in culture supernatants of not infected (n = 4) or SARS-CoV-2 infected macrophages and foam cells with (n = 4) or without (n = 8) NRP1-blocking treatment (EG00229 trifluoroacetate) at 24 hpi. Data are presented as mean values ±s.e.m. One-way ANOVA with post-hoc Tukey’s test for multiple comparisons was performed.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Tables
Supplementary Tables 1–5.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Eberhardt, N., Noval, M.G., Kaur, R. et al. SARS-CoV-2 infection triggers pro-atherogenic inflammatory responses in human coronary vessels. Nat Cardiovasc Res 2, 899–916 (2023). https://doi.org/10.1038/s44161-023-00336-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44161-023-00336-5
- Springer Nature Limited
This article is cited by
-
Mechanisms of endothelial activation, hypercoagulation and thrombosis in COVID-19: a link with diabetes mellitus
Cardiovascular Diabetology (2024)
-
Three-year outcomes of post-acute sequelae of COVID-19
Nature Medicine (2024)
-
Identification of the needs of individuals affected by COVID-19
Communications Medicine (2024)
-
A COVID-19 specific multiparametric and ECG-based score for the prediction of in-hospital mortality: ELCOVID score
Internal and Emergency Medicine (2024)
-
SARS-CoV-2 infection boosts inflammation in atherosclerotic plaques
Nature Cardiovascular Research (2023)