Abstract
RAB35 is a multifunctional small GTPase that regulates endocytic recycling, cytoskeletal rearrangement, and cytokinesis. However, its physiological functions in mammalian development remain unclear. Here, we generated Rab35-knockout mice and found that RAB35 is essential for early embryogenesis. Interestingly, brain-specific Rab35-knockout mice displayed severe defects in hippocampal lamination owing to impaired distribution of pyramidal neurons, although defects in cerebral cortex formation were not evident. In addition, Rab35-knockout mice exhibited defects in spatial memory and anxiety-related behaviors. Quantitative proteomics indicated that the loss of RAB35 significantly affected the levels of other RAB proteins associated with endocytic trafficking, as well as some neural cell adhesion molecules, such as contactin-2. Collectively, our findings revealed that RAB35 is required for precise neuronal distribution in the develo** hippocampus by regulating the expression of cell adhesion molecules, thereby influencing spatial memory.
Similar content being viewed by others
Introduction
The RAB proteins form the largest family of small GTPases that regulate intracellular membrane trafficking in eukaryotic cells1. Particularly, RAB35 is a highly conserved RAB protein in metazoans and is localized in the recycling endosome and plasma membrane. RAB35 was originally discovered as a regulator of endocytic recycling required for cytokinesis in HeLa cells2,3. In Caenorhabditis elegans, RAB35 controls the endocytic recycling of yolk receptors essential for oocyte growth and cell death4,5,6. Drosophila RAB35 functions in actin bundling, phagocytosis, neurotransmitter release, and spermatogenesis7,8,9,10. In mammals, RAB35 regulates several membrane trafficking pathways, including endosome-to-Golgi retrograde transport, phagocytosis, exosome secretion, and autophagy, in addition to its primary role in the recycling pathway8,11,12,13. Mammalian RAB35 also plays a role in multiple cellular processes associated with dynamic membrane changes, such as cytokinesis, immunological synapse formation, myoblast fusion, cilium length, and cell migration, in many kinds of culture cells and primary cells2,14,15,16,17,18. Additionally, previous studies in Drosophila and neuronal cell lines have reported several functions of RAB35 in the nervous system, including neurite outgrowth, axon elongation, exosome secretion, neurotransmitter release, oligodendrocyte differentiation, and synaptic vesicle turnover9,11,19,20,21,22. In fact, associations between RAB35 and neurodegenerative diseases, such as Parkinson’s disease and Alzheimer’s disease, have recently emerged23,24. Recently, RAB35 has also been identified as an oncogenic RAB that promotes cell proliferation25. However, the physiological importance of RAB35 in mammalian development remains largely unknown.
The formation of the mammalian brain is a highly orchestrated process that involves the generation, differentiation, migration, and maturation of neurons. Neuronal migration is an essential process to ensure the proper positioning of neurons and for the organization of multilayered structures, notably the cerebral cortex and hippocampus26. The impairment of this process has an adverse effect on neural networks and brain architecture, causing several neurological disorders, including lissencephaly, epilepsy, and schizophrenia27,28,29. The molecular mechanism associated with the formation of laminated structures has been primarily investigated in the cerebral cortex. Knockdown experiments based on in utero electroporation have revealed that RAB proteins related to endocytosis, such as RAB5, RAB7, RAB11, RAB18, and RAB23, regulate cortical neuronal migration in mice by controlling the traffic of N-cadherin during the distinct steps of endocytosis30,31,32. Although the formation process of the hippocampus is believed to be similar, its developmental mechanism, including the role of RAB proteins, is less well-understood.
To reveal the physiological function of RAB35 in mammals, here, we generated systemic Rab35-knockout mice and found that RAB35 is essential during early mouse embryogenesis. We also generated central nervous system (CNS)-specific Rab35-knockout mice and identified RAB35 as a requirement for proper hippocampal development. Our findings suggest that RAB35 plays an essential role in the proper positioning of pyramidal neurons in the hippocampus by regulating the expression of surface cell adhesion molecules, as well as being involved in appropriate spatial memory formation.
Results
RAB35 is essential for early mouse development
To investigate the physiological function of RAB35 in mammalian development, we generated Rab35-knockout mice using an ES cell line (Supplementary Fig. 1a). The disruption of the Rab35 gene was validated using Southern blotting and PCR (Supplementary Fig. 1b, c). Heterozygous-targeted mice (Rab35geo/+ or −/+) were healthy and fertile. To obtain the homozygous knockout mice (Rab35geo/geo and Rab35−/−), we intercrossed the heterozygous knockout and found that no offspring were either Rab35geo/geo or Rab35−/−, indicating that Rab35 null mice are embryonic lethal. At embryonic day (E) 15.5 and E11.5, no Rab35 null embryos were obtained. At E9.5, 9 (32%) embryos were Rab35 +/+; 13 (46%) embryos were Rab35−/+; and 6 (21%) embryos were Rab35−/− (Supplementary Fig. 1d). These results indicate that Rab35 is essential for early mouse development.
RAB35 protein expression gradually increases during early brain development
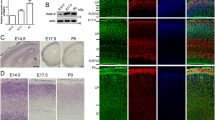
Previous studies in Drosophila and neuronal cell lines have implied that RAB35 plays an important role in diverse neural functions, including neurotransmitter release, neurite elongation, oligodendrocyte differentiation, and exosome secretion9,11,15,20. We thus focused on the in vivo function of RAB35 in brain development. To this end, we first determined the expression profiles of RAB35 in the mouse brain. RAB35 protein levels gradually increased from E15 and peaked at postnatal day (P) 14 in the develo** brain (Fig. 1a, b). Considering the whole brain, the RAB35 protein was expressed in the hippocampus at E15.5, with expression gradually increasing during development (Fig. 1c, d). LacZ staining detected RAB35 expression in several brain subregions, including the cerebral cortex, hippocampus, and thalamus at E15.5 (Fig. 1e). In the P1 brain, expression was more notable in the hippocampus and over a wide area of the cerebral cortex. In addition, RAB35 expression was identified in the olfactory bulb and cerebellum of adult mouse brains (Fig. 1f, g). Furthermore, we examined the cell types expressing RAB35 in adult mouse brains by immunostaining for β-galactosidase using either the mature neuronal marker NeuN or the astrocyte marker glial fibrillary acidic protein (GFAP). Maximum intensity projection images revealed β-galactosidase puncta in NeuN-positive and GFAP-positive cells, suggesting that RAB35 is expressed in neurons and astrocytes (Fig. 1h, i).
a RAB35 protein levels in the whole mouse brain during the embryonic (E15.5 and E17.5) and postnatal (P2, P6, P14, and P21) periods. b Quantification of RAB35 protein levels in the whole mouse brain. Band intensities of RAB35 were normalized against those of actin (n = 3 per the indicated stage). c RAB35 protein levels in the hippocampus during the embryonic (E15.5 and E17.5) and postnatal (P2, P6, P14, and P21) periods. d Quantification of RAB35 protein levels in the mouse hippocampus (n = 3 per stage). e X-gal staining of sagittal sections from E15 and P1 Rab35geo/+ mouse brains. Scale bars, 200 μm. f RAB35 protein levels in 3-month-old mouse brain subregions. Ob olfactory bulb, Cx cerebral cortex, Hp hippocampus, Th thalamus, Cb cerebellum. g X-gal staining of a sagittal section of a 3-month-old Rab35geo/+ mouse brain. Scale bars, 500 μm. h, i Representative maximum intensity projection images of 4-month-old Rab35geo/+ mouse brain stained for β-galactosidase (green), NeuN or GFAP (magenta), and DAPI (blue). Scale bar; upper panel, 50 μm; lower panel, 20 μm. Data represent mean ± SEM.
To investigate the physiological importance of RAB35 in neural development, we generated CNS-specific Rab35-knockout mice by crossing mice harboring the Rab35flox allele with transgenic mice expressing Cre recombinase under the control of the Nestin promoter (Nestin-Cre)33(Supplementary Fig. 1a). Although RAB35 protein expression was detectable in the mouse brain at E13.5 (Fig. 2a), it was undetectable in the whole brain of E13.5 Rab35flox/flox; Nestin-Cre embryos and in the cerebral cortex and the hippocampus of 4-month-old Rab35flox/flox; Nestin-Cre mice (Fig. 2a, b). CNS-specific Rab35 conditional knockout (cKO) mice were born at the Mendelian frequency and were viable for more than 1 year. Male and female Rab35 cKO mice gradually showed lower body weight than the control Rab35flox/+; Nestin-Cre mice. (Supplementary Fig. 1e, f).
a, b Western blot analysis of E13.5 (a) and 4-month-old (b) Rab35flox/flox, Rab35flox/+; Nestin-Cre, and Rab35flox/flox; Nestin-Cre mouse brain lysates. c–f Open field test in control (n = 13) and Rab35 cKO (n = 17) mice. c Typical traces of the horizontal locomotor activity. d Distance traveled per minute. Two-way repeated-measures ANOVA; interaction effect, F(14, 392) = 1.094, p = 0.3612. e Total distance traveled. Mann–Whitney test, p = 0.3. f Stay time in the center area. Mann–Whitney U-test, p < 0.001. g–j Elevated plus maze in control (n = 20) and Rab35 cKO (n = 22) mice. g Cumulative percentage of mice falling from open arms. h Typical traces in the elevated plus maze test. i Stay time in open arms. Unpaired Student’s t-test, p = 0.0054. j Stay time in the closed arms. Mann–Whitney U-test, p = 0.0034. k Rotarod test in control (n = 23) and Rab35 cKO (n = 20) mice. Two-way repeated-measures ANOVA with Bonferroni correction; interaction effect, F(5, 205) = 3.009, p = 0.0121; trial 3, p = 0.0076; trial 4, p < 0.0001; trial 5, p = 0.0059; trial 6, p = 0.0008. l, m Barnes maze tests in control (n = 16) and Rab35 cKO (n = 19) mice. l Latency to reach the target hole. Each point represents the mean of three trials. Two-way repeated-measures ANOVA with Bonferroni correction; interaction effect, F(3, 99) = 1.706, p = 0.1706; day 1, p = 0.0261; day 4, p = 0.0030. m Time spent around the target hole during a 90 s period. Mann–Whitney U-test, p = 0.0136. n Alternation percentages in Y maze test of control (n = 15) and Rab35 cKO (n = 18) mice. Unpaired Student’s t-test, p = 0.01454. Data represent mean ± SEM or box and whisker plots; box plot shows 25th and 75th percentiles (box), median (horizontal bar), and minimum to maximum (whiskers); n.s. not significant (p > 0.05); *p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001.
CNS-specific Rab35 cKO mice exhibit reduced anxiety-related behaviors and motor function
To evaluate the impact of RAB35 deficiency on mouse behavior, we performed behavioral tests. In the open field test, which measures spontaneous motor activity and anxiety-related behavior in a novel environment, Rab35 cKO mice exhibited spontaneous movement with travel times similar to the control mice (Fig. 2c–e), indicating that their horizontal locomotor activities were comparable. Meanwhile, the time spent in the center of the open field apparatus significantly increased in Rab35 cKO mice (Fig. 2c, f). We then performed the elevated plus maze test to assess anxiety-related behaviors. Although 13 of the 22 Rab35 cKO mice dropped from the open arms during the test (Fig. 2g), we evaluated the anxiety levels of nine mice that completed the test without falling. Rab35 cKO mice spent significantly more time in the open arms and less time in the closed arms than control mice (Fig. 2h–j). These results indicated that Rab35 cKO mice exhibit reduced anxiety-related behaviors.
In the accelerated rotarod test, which is used to assess motor function, control mice increased the time spent on the rotating rod after several trials, with most control mice eventually spending more than 150 s on a rod. In contrast, Rab35 cKO mice exhibited a significantly reduced latency to fall (Fig. 2k). Taken together, these results suggest that RAB35 deficiency does not affect horizontal locomotor activity but causes defects in anxiety-related behaviors and motor function.
Rab35 cKO mice show a defect in spatial learning and memory
We further assessed spatial learning and memory using the Barnes maze test (Fig. 2l, m). The Barnes maze consists of a circular table with 16 holes around the perimeter and visual cues around the table. Of the 16 holes, only one contains an escape box underneath, considered the target. This test consists of several phases, including the acquisition phase and the probe test. During the acquisition phase, the spatial learning ability of each mouse was evaluated by measuring the time required to reach the target. As expected, the control mice learned the location of the target hole to avoid bright room lighting, with reduced time required to reach the target in each trial. Rab35 cKO mice spent significantly more time reaching the target than control mice on day 4 (Fig. 2l). The probe test was conducted one day after the last acquisition trial to examine spatial memory acquisition. We removed the escape box from the table and measured the time spent in the proximity of the target to determine whether the mice had memorized the target hole position. As shown in Fig. 2m, Rab35 cKO mice spent slightly but significantly less time around the correct target hole than control mice. To analyze the potential effects of RAB35 on short-term memory, mice were examined in a Y maze test. The percentage of spontaneous alternation in Rab35 cKO mice was comparable with that in control mice (Fig. 2n). These results suggest that Rab35 cKO mice exhibit deficits in spatial memory but not in working memory.
The laminated structure of the cerebral cortex is preserved in Rab35 cKO mice
Next, we investigated whether the loss of RAB35 affects brain architecture. The mammalian cerebral cortex has a six-layered structure. During cortical development, excitatory neurons are generated from radial glial cells in the ventricular side34,35. Newly born neurons radially migrate from the apical ventricular side toward the basal pial surface and stop migrating just beneath the marginal zone. Therefore, early-born neurons form the deep layer (layers VI and V), whereas late-born neurons pass through the deep layer and settle in more superficial layers (layers IV–II); hence, the proper six-layered structure of the cerebral cortex is formed in a birthdate-dependent inside-out manner36,37. Cortical neurons are segregated into cell layers with neurons formed around the same period and expressing layer-specific transcription factors, including chicken ovalbumin upstream promoter transcription factor-interacting proteins 2 (Ctip2; a marker for layer V) and cut homeobox 1 (Cux1; a marker for layer II–IV), to differentiate and establish layer-dependent neural connections.
The dorsal view or brain weight of Rab35 cKO mice was comparable to that of control mice (Fig. 3a, b). In Rab35 cKO mice, the thickness of the cerebral cortex and the relative thickness of the Cux1-positive superficial layer (layers II–IV), as well as that of the Ctip2-positive deep layer (layer V), were comparable with those of control mice at P0, the period when most of the cortical excitatory neurons reach their layer (Fig. 3c–f). We further evaluated neuronal migration in the cerebral cortex using the 5-ethynyl-2-deoxyuridine (EdU) labeling birthdate analysis (Fig. 3g, h). EdU was intraperitoneally injected into pregnant mice at E14.5, and the brains of the pups were collected at P12. The cerebral cortex was divided into 10 equal bins from the ventricular to pial surfaces, and the proportion of EdU-positive cells in each bin was quantified. RAB35-deficient cortical neurons labeled with EdU did not show significant migration defects. Immunostaining for NeuN revealed no significant difference in the distribution of NeuN-positive cells between the cortices of control and Rab35 cKO adult mice (Fig. 3i). The thickness of the cerebral cortex and the number of NeuN-positive cells in the 300-μm wide cortices were also shown to be maintained in Rab35 cKO mice (Fig. 3j, k). Thus, these results indicate that the laminated architecture is preserved in the cerebral cortex of RAB35-deficient mice.
a Dorsal view of 3-month-old control and Rab35 cKO mice brains. Scale bar, 2 mm. b Brain weights of 3-month-old control (n = 7) and Rab35 cKO (n = 8) male mice. Mann–Whitney U-test, p = 0.8409. c Representative images of P0 cortices of the control and Rab35 cKO mice immunostained for Cux1 (magenta), Ctip2 (green), and DAPI (blue). Scale bar, 100 μm. d–f Quantification of the cortical (d), relative Cux1-positive (e), and Ctip2-positive (f) layer thicknesses of P0 control (n = 4) and Rab35 cKO (n = 4) mice. Cux1-positive and Ctip2-positive layer thicknesses were measured and normalized against total cortical thickness. Mann–Whitney U- test; d, p = 0.6857; e, p >0.99; f, p = 0.2. g Representative EdU labeling birthdate analysis images of P12 cortex samples stained with EdU (magenta) and DAPI (blue). Scale bar, 100 μm. h Proportion of EdU-positive neurons in P12 cortices of control (n = 4) and Rab35 cKO (n = 4) mice. Two-way repeated-measures ANOVA with Bonferroni correction; interaction effect, F(9, 54) = 0.4991, p = 0.8687. i Immunohistochemistry of sagittal sections of cerebral cortices of 4-month-old control and Rab35 cKO mice using an anti-NeuN antibody. Scale bars, 100 μm. j, k Quantitative analysis of the cortical thickness (j) and the number of NeuN-positive cells in the 300-μm width cerebral cortex (k) of the control (n = 5) and Rab35 cKO (n = 5) mice. Unpaired Student’s t-test; j, p = 0.6928; k, p = 0.5375. Data represent mean ± SEM or box and whisker plots; box plot shows 25th and 75th percentiles (box), median (horizontal bar), and minimum to maximum (whiskers); n.s., not significant (p > 0.05).
Loss of RAB35 induces heterotopic positioning of neurons in the hippocampus
In contrast with the cerebral cortex, we found that the laminated structure of the hippocampus was severely disorganized in Rab35 cKO mice (Fig. 4a). The hippocampus contains a tightly packed pyramidal cell layer, also called stratum pyramidale (SP), of the Ammon’s horn (CA1, CA2, and CA3 regions) and a granule cell layer (GCL) of the dentate gyrus (DG) as shown in the control mice. In the Rab35 cKO hippocampus, however, the SP in CA1 was remarkably fractured into stratum oriens (SO), and the pyramidal cells in CA3 were distributed more extensively. Ectopic pyramidal neurons were observed in the ventral hippocampal CA1 region (Supplementary Fig. 2a). Based on quantitative bin analysis, Rab35 cKO mice exhibited significant changes in pyramidal cell distribution in CA1 and CA3 (Fig. 4b, c). The number of NeuN-positive neurons in either the RAB35-deficient CA1 or CA3 regions were comparable to those in controls (Supplementary Fig. 2b, c). Additionally, the granule cells in the DG dispersed towards the molecular layer with an increased cell number in Rab35 cKO mice (Fig. 4d and Supplementary Fig. 2d). The CA1 neurons positive for Ctip2, here used as a postmitotic neuronal marker, presented a divided cell layer at P6, the period when most newly born hippocampal excitatory neurons arrive at their final position (Supplementary Fig. 2e), suggesting that hippocampal neurons ectopically position during the developmental stage.
a Immunohistochemistry of sagittal sections of hippocampi of 4-month-old control and Rab35 cKO mice using an anti-NeuN antibody. SP: stratum pyramidale, SO: stratum oriens. Scale bars, 200μm. b–d Distribution of NeuN-positive cells in the hippocampal CA1 (b), CA3 (c), and DG (d) regions of control (n = 4) and Rab35 cKO (n = 4) mice. Two-way repeated-measures ANOVA with Bonferroni correction; b interaction effect, F(4, 32) = 4.161, p = 0.008; c interaction effect, F(4, 32) = 3.264, p = 0.0236; d interaction effect, F(4, 32) = 25.9, p < 0.0001. e Representative images of P12 hippocampi stained with EdU (magenta) and DAPI (blue). Scale bar, 100 μm. f Proportion of EdU-positive neurons in the P12 hippocampi of control (n = 4) and Rab35 cKO (n = 4) mice. Two-way repeated-measures ANOVA with Bonferroni correction; interaction effect, F(4, 24) = 3.712, p = 0.0173. g Representative images of DIV 3 hippocampal primary neurons stained for p-NF (green) with rhodamine-phalloidin (magenta) and DAPI (blue). Scale bar, 100 μm. h, i Quantification of the rhodamine-phalloidin-positive total neurite length (h) and p-NF-positive axon length (i) of the control and Rab35-deficient primary neurons at DIV 3. At least 50 neurons from four (control) and three (Rab35 cKO) different cultures were measured. Unpaired Student’s t-test; h, p = 0.8204; i, p = 0.8832. j Representative ventral hippocampal commissure (VHC) images of 3-month-old control and Rab35 cKO mice (bregma −0.35 mm) by Klüver–Barrera staining. Scale bar, 200 μm. k VHC thickness in control (n = 6) and Rab35 cKO (n = 7) mice. Unpaired Student’s t-test, p = 0.0695. Data represent mean ± SEM; n.s. not significant (p > 0.05); *p < 0.05; **p < 0.01; and ***p < 0.001.
Loss of RAB35 leads to an impaired distribution of hippocampal pyramidal neurons
Since the rodent hippocampal pyramidal neurons in the CA1–CA3 regions are generated from a neuroepithelium dorsally shared with the neocortex, the laminated structures in the CA regions of the hippocampus are considered to form similar patterns to those in the cerebral cortex27,38. During the development of the cerebral cortex and the hippocampus, excitatory neurons are generated from two major types of progenitor cells: radial glial cells in the ventricular zone (VZ) and intermediate progenitors in the subventricular zone (SVZ)34,35. Radial glial cells produce intermediate progenitors or neurons in the VZ via asymmetric division, whereas intermediate progenitors migrate and accumulate in the subventricular zone (SVZ) just above the VZ. The formation of the laminated structures in the hippocampus primarily depends on the highly coordinated production and migration of neurons from the proliferative zones. The spatiotemporal control of neural progenitor cell proliferation is important for the production and positioning of neurons. In addition to proliferative potential, the morphology of radial glial cells also plays an important role in neuronal positions because newborn neurons use radial processes as scaffolds to migrate toward the final position in primitive SP.
We then investigated the functional and morphological integrity of the neural progenitors in the Rab35 cKO hippocampus. The distributions of the paired box protein 6 (Pax6)-positive radial glial cells and T-box brain protein 2 (Tbr2)-positive intermediate progenitors were similar in the control and Rab35 cKO CA regions at E15.5 (Supplementary Fig. 2f, g). The number of both progenitors was comparable in control and Rab35 cKO mice (Supplementary Fig. 2h, i). Immunostaining for phosphorylated histone H3 (pHH3), a marker for cells undergoing mitosis, showed that the number of pHH3-positive cells in the Rab35 cKO CA region did not change significantly compared with the control mice (Supplementary Fig. 2j, k). Similar results were obtained in DG (Supplementary Fig. 2l–n). These results indicate that the conditional deletion of Rab35 using Nestin-Cre transgenic mice does not affect the spatial pattern and proliferation potency of neural progenitors in the hippocampus. Furthermore, we performed immunostaining using an antibody for Nestin, an intermediate filament protein in radial glial cells, to observe the morphology of the radial processes. Nestin-positive radial glial processes appeared to be normally organized in the sagittal sections of E15.5 Rab35 cKO hippocampus (Supplementary Fig. 2o). From these data, the function and shape of the hippocampal neural progenitor cells are maintained in Rab35 cKO mice.
Given that the loss of RAB35 did not impact neural progenitor cells, we examined whether malformation of the hippocampal layer could be attributed to defective migration or positioning of neurons during development using EdU labeling birthdate analysis (Fig. 4e, f). The hippocampal CA1 region, from the edge of the apical surface to 400 μm, was divided into five equal bins, and the proportion of EdU-positive cells in each bin was quantified. In control mice, over 90% of EdU-positive neurons reached bins 2 and 3, whereas, in Rab35 cKO mice, the proportion of EdU-positive neurons in bins 2 and 3 decreased with an increase in bins 1 and 4 (Fig. 4f), thereby suggesting that neuronal cells could migrate, but were not distributed to the precise location. These findings indicate that RAB35 is required for the precise distribution of pyramidal cells in the develo** hippocampus.
Rab35-deficient hippocampal neurons exhibit superficially normal neuritogenesis
Based on previous studies that RAB35 knockdown suppresses the elongation of neurites in PC12 cells and axons in rat hippocampal primary neurons and the overexpression of wild-type or constitutively active mutant RAB35 enhances the elongation, we examined neuritogenesis in primary neurons harvested from the Rab35 cKO mice. Hippocampal primary neurons cultured for days in vitro (DIV) 3 were subjected to immunostaining for an axon marker phosphorylated neurofilament heavy chain (pNF-H) and rhodamine-phalloidin (Fig. 4g). The lengths of both the phalloidin-positive total neurite and pNF-H-positive axon in RAB35-deficient primary neurons were comparable with those in control neurons (Fig. 4h, i), indicating that primary neurons derived from the Rab35 cKO hippocampus sustain sufficient potential for neuritogenesis. To evaluate the role of RAB35 on axon elongation in vivo, we examined the thickness of the ventral hippocampal commissure (VHC) (Fig. 4j, k). Rab35 cKO mice tended to exhibit a relatively reduced VHC thickness when compared with that of control mice; however, this difference was not statistically significant. These findings suggest that neuritogenesis is not markedly impaired despite the absence of RAB35.
RAB35 is required for proper cell surface expression of neural cell adhesion molecules (NCAMs) in hippocampal neurons
To gain insight into the molecular mechanism by which RAB35 contributes to neuronal migration, we assessed the differential protein expression in the hippocampus from P0 in control and Rab35 cKO mice using a multiplexed isobaric tandem mass tag (TMT) labeling and LC-MS/MS analysis. In the P0 hippocampus, neurogenesis passed the peak, and most pyramidal neurons migrated toward their position. Among the 7495 proteins identified and quantified using P0 hippocampal proteomics, 193 were significantly upregulated or downregulated when the threshold was set to p < 0.01 (Fig. 5a and Supplementary Data 1). Consistent with the western blot analysis (Fig. 2a, b), we confirmed that the RAB35 protein was markedly decreased. This dataset included 37 proteins related to membrane traffic, especially the recycling and endocytic pathways, and 35 of 37 proteins, including RAB11B, RAB4A, Snx3, Atg9a, Vamp4, and RAB7 were decreased in the Rab35 cKO hippocampus (Fig. 5b and Supplementary Table 1), suggesting that the loss of RAB35 might disturb the recycling and endocytic pathways.
a Volcano plot of the TMT-based quantitative proteomes identifying the dysregulated proteins in Rab35 cKO hippocampus in comparison with the control hippocampus (n = 5 mice per genotype). b Number of proteins identified as significantly dysregulated and as either membrane traffic-related or neuronal migration-related. c, d Western blot analysis of control and Rab35 cKO P0 hippocampi using anti-contactin-2, anti-CHL1, and anti-actin antibodies. e, f Quantification of contactin-2 (c) and CHL1 (d) protein levels in control and Rab35 cKO P0 hippocampi. Band intensities of the indicated proteins were normalized to those of actin (n = 9 mice per genotype). Unpaired Student’s t-test; e, p = 0.0153; f, p = 0.0095. g, h Levels of contactin-2 (g) and CHL1 (h) were quantified by targeted MS using the PRM method (n = 5 mice per genotype). Unpaired Student’s t-test; g p = 0.0053; h p = 0.0229. i Western blot analysis of control and Rab35 cKO P0 hippocampus using anti-N-cadherin and anti-actin antibodies. j Quantification of N-cadherin protein levels in the control and Rab35 cKO P0 hippocampus (n = 9 mice per genotypes). Unpaired Student’s t-test, p = 0.9020. k Representative images of DIV 2 hippocampal primary neurons stained for contactin-2 (green), rhodamine-phalloidin (magenta) and DAPI (blue). Scale bar, 20 μm. l Quantification of contactin-2 intensity at the somatic plasma membrane in control (n = 4) and Rab35-deficient (n = 4) cells. Thirty neurons from four different cultures per genotype were analyzed. Mann–Whitney U-test, p = 0.0286. Data represent the mean ± SEM; n.s. not significant (p > 0.05); *p < 0.05; **p < 0.01.
In the dataset, we found that 25 neuronal migration-related proteins are affected by the loss of RAB35. Among these, contactin-2/transiently expressed the axonal surface glycoprotein-1 (TAG-1) and the close homolog of L1 (CHL1) was significantly more abundant in Rab35 cKO hippocampus than in control mice (Fig. 5a and Supplementary Table 2). Contactin-2 and CHL1 are neural cell adhesion molecules (NCAMs) of the immunoglobulin superfamily (IgSF) belonging to the contactin and L1 subfamilies, respectively. Contactin-2 and CHL1 are present on the cell surface and mediate cell-cell and cell-substratum adhesions via the homophilic and/or heterophilic interactions with other cell adhesion molecules and the extracellular matrix to play a role in axon growth and in guiding migrating neurons in the develo** brain. We confirmed that the protein levels of both contactin-2 and CHL1 were significantly increased in P0 Rab35 cKO hippocampus using western blot analysis and parallel reaction monitoring (PRM)-based targeted mass spectrometry (Fig. 5c–h). The protein levels of other cell adhesion molecules, including the cadherin family N-cadherin, were unchanged in control and Rab35 KO mice (Fig. 5i, j). In DIV 2 hippocampal neurons isolated from E18 embryos, contactin-2 was shown to be localized at the plasma membrane of the soma and neurites (Fig. 5k). Moreover, the fluorescence intensity of contactin-2 at the somatic plasma membrane of RAB35-deficient neurons was significantly higher when compared to control neurons (Fig. 5l). Exogenously expressed GFP-RAB35 tended to reduce the level of the contactin-2 signal on the somatic plasma membrane when compared with that in control cells; however, the difference was not statistically significant (Supplementary Fig. 2p). Thus, these results suggest that RAB35 ablation affects protein homeostasis in the develo** hippocampus, resulting in increased contactin-2 and CHL1 levels, most likely due to a defect in internalization from the cell surface and/or recycling back to the plasma membrane.
Discussion
RAB35 plays an important role in diverse cellular processes associated with dynamic membrane changes, such as endocytic recycling, cytokinesis, cell migration, and neurite outgrowth. However, the physiological function of mammalian RAB35 is yet to be investigated. This study, involving systemic and CNS-specific Rab35-knockout mice, identified RAB35 as an essential component in hippocampal development by regulating neuronal positioning properly.
Systemic Rab35 knockout caused embryonic lethality until E11.5, indicating that RAB35 is essential for embryonic development. Rab35 homozygous knockout embryos were obtained at E9.5, at the expected Mendelian frequency (Supplementary Fig. 1), suggesting that Rab35-knockout embryos were implanted but failed to develop. After implantation, gastrulation forms essential layers, including the ectoderm, mesoderm, and endoderm. Visceral endoderm cells exhibit high endocytic activity, supply maternal nutrients, and control signaling cascades for embryonic growth. Interestingly, RAB7 deficiency perturbs endocytic flow in visceral endoderm cells and causes reduced levels of intracellular amino acids, along with dysregulated cellular signaling, resulting in a lethal embryonic phenotype at E739,40. One possible explanation for the essential nature of RAB35 during development is that Rab35-knockout embryos exhibit defects in recycling pathways in the visceral endoderm. Additional investigations are required to establish the role of RAB35 in mouse embryogenesis. Furthermore, we also generated CNS-specific Rab35 cKO mice to examine the role of RAB35 in brain development. CNS-specific Rab35 cKO mice had comparable brain weights to control mice and preserved the laminated structure of the cerebral cortex (Fig. 3). Although RAB35 is involved in neuritogenesis, neurons from Rab35 cKO mice showed elongated axons in vitro and in vivo (Fig. 4). This discrepancy may arise because of differences in the cell lines/tissues used or differences between chronic and acute depletion of RAB35. Alternatively, other RAB proteins may compensate for the defects caused by the loss of RAB35 during early brain development.
In contrast, RAB35-deficient hippocampi showed a disorganized cell layer through the CA and DG regions (Fig. 4). Hippocampal lamination is formed under the strict spatiotemporal control of the neuronal production and the migration of neurons from the proliferative zones. The proliferative potential and morphology of radial glial cells were normal in Rab35 cKO mice (Supplementary Fig. 2). These findings indicate that neither defective neuronal proliferation from radial glial cells nor radial processes used as a scaffold for migrating neurons are the primary causes for the malpositioning of mutant neurons. In the birthdate analysis, Rab35 cKO neurons migrated but failed to migrate to precise locations. These data indicate that RAB35 is required to precisely distribute pyramidal cells in the develo** hippocampus.
Although several RAB proteins associated with endocytosis (RAB5, 7, 11, 18, 23) have been reported to play important roles in the formation of the six-layered structure in the cerebral cortex by regulating distinct steps of neuronal migration30,31,32, the involvement of these RAB proteins in hippocampal lamination remains unknown. Interestingly, RAB35-deficient mice showed a superficially normal cerebral cortex structure but exhibited defective hippocampal lamination, suggesting that RAB35 contributes more predominantly to proper neuronal positioning in the hippocampus than in the cerebral cortex.
How does the loss of RAB35 widely perturb membrane trafficking in hippocampal neurons? One possible explanation is that the loss of RAB35 alters phosphoinositide (PI) homeostasis and leads to defects in endocytic trafficking. RAB35 is known to regulate PI homeostasis, especially PtdIns(4,5)P2 and PtdIns3P/ PtdIns(3,5)P2, in coordination with its effectors, ACAP2 and myotubularin phosphatases, respectively. ACAP2 is a GTPase-activating protein (GAP) for Arf6, which activates its effector PIP5K to synthesize PtdIns(4,5)P2. Activated RAB35 localizes to Arf6-positive compartments and then recruits ACAP2 to inactivate Arf6 in order to terminate PtdIns(4,5)P2 synthesis3,41,42. In contrast, recent studies have reported that RAB35 controls the turnover of PtdIns3P and PtdIns(3,5)P2 by binding to myotubularin-related protein 13- and MTMR2-containing lipid phosphatase complexes, which hydrolyze the 3-phosphate from PtdIns3P and PtdIns(3,5)P2 located on endosomes. Thus, the lack of RAB35 could provoke a local increase in PtdIns(4,5)P2, PtdIns3P, and PtdIns(3,5)P2, thereby perturbing the recycling and endocytic pathways. Indeed, the results of the proteomics analysis revealed that the levels of various endocytic regulators, such as Snx3 and Snx27, both of which bind to PtdIns3P and PIP5k1c, were significantly reduced in RAB35-deficient hippocampal lysates (Fig. 5 and Supplementary Table 1).
We also found that IgSF cell surface proteins, namely contactin-2 and CHL1, increased in the develo** Rab35 cKO hippocampus. It has been reported that CHL1 plays a role in neuronal migration in the cerebellum and caudal cerebral cortex43,44,45. During cerebellar development, the L1 family protein CHL1 controls neuronal migration via heterophilic trans-interactions of CHL1 with vitronectin and integrins. Furthermore, the cytoplasmic regions of CHL1 interacted with ezrin and ankyrin to participate in actin dynamics46,47. The increase or decrease in the level of another L1 family protein retarded the migration of cortical neurons48,49. Contactin-2 has also been reported to be involved in the migration of cortical interneurons and caudal medulla neurons50,51. Contactin-2 bound to L1 via cis-interaction and enabled L1 to recruit Ankyrin52. Given that RAB35 is required for the endocytosis and recycling of cell surface proteins, loss of RAB35 could disturb the recycling and targeting of these molecules to the appropriate locations for neuronal migration, thereby resulting in the disorderly distribution of pyramidal neurons and granule cells in the hippocampus. Moreover, RAB35 deficiency may impact intracellular actin dynamics. Alternatively, the loss of RAB35 may cause aberrant local accumulation of PI derivatives on the membrane compartments, thereby disturbing the endocytic trafficking of cell surface proteins.
Behavioral tests revealed that CNS-specific Rab35 cKO mice have defects in a broad range of behaviors, such as anxiety-related behaviors and motor function, as well as spatial memory. (Fig. 2). The dorsal hippocampus is responsible for spatial learning and memory, whereas the ventral hippocampus functions as part of the anxiety circuitry. Thus, reduced spatial memory performance in Rab35 cKO mice could be associated with disorganized hippocampal lamination. Several brain regions, including the amygdala, striatum, ventral hippocampus, and prefrontal cortex, modulate anxiety-related behaviors. Our data suggest that disrupted layer formation in the ventral hippocampus of Rab35 cKO mice may affect anxiety-like behaviors. Since RAB35 is also expressed in other brain regions, including the striatum and prefrontal cortex, the loss of RAB35 in such regions may contribute to reduced anxiety-related behaviors in Rab35 cKO mice. Therefore, further studies are needed to understand the physiological roles of RAB35 in brain development and function.
Recent studies have suggested associations between RAB35 and neurodegenerative diseases. Our proteomics data showed a significant decrease in Alzheimer’s disease-related factors LRP1 and presenilin in RAB35-deficient hippocampus. Further characterization of the physiological and pathological functions of RAB35 will provide a better understanding of the molecular pathology of neurodegenerative diseases.
Methods
Generation of Rab35-knockout mice
Rab35-knockout mice were generated as described previously53. Rab35-knockout ES clones were purchased from EUCOMM, with the SA-IRES-βgeo-polyA sequence inserted into the second intron of the Rab35 gene. The targeted ES clones were identified using Southern blot analysis and injected into the BALB/c embryos to generate the Rab35geo/+ mice. The mice were back-crossed more than six generations to the C57BL/6 background. To generate the Rab35flox/+ and Rab35−/+ mice, the Rab35geo/+ mice were crossed with Act-Flp-e and CAG-Cre transgenic mice (Jackson Laboratory), respectively. To generate the CNS-specific Rab35-knockout mice, Rab35flox/+ mice were crossed with Nestin-Cre transgenic mice33. The genotypes of each mouse line were confirmed through PCR using the following primers: primer 1 (5′ arm), 5′-ACACTTCACATGGCTCTCTGGTCC-3′; primer 2 (3′ arm), 5′-CTCTAGCAGACCCACAATGCGAGC-3′; primer 3 (LAR3), 5′-CAACGGGTTCTTCTGTTAGTCC-3′; and primer 4 (40 R), 5′-TGAACTGATGGCGAGCTCAGACC-3′. The cre gene was identified through PCR using the following primers: forward, 5′-AGGTTCGTTCACTCATGGA-3′; and reverse, 5′-TCGACCAGTTTAGTTACCC-3′. All animal procedures were approved and performed in accordance with the guidelines of the Animal Care and Experimentation Committee of Gunma University, Japan (approval no. 19-001). Mice were bred at the Bioresource Center of Gunma University, Graduate School of Medicine, Japan.
Antibodies
The following primary antibodies were used for immunoblotting: 1:1000 RAB35 (rabbit; Cell Signaling, 9690 S), 1:10,000 Actin [C4] (mouse; Merck-Millipore, MAB1501), 1:10,000 GAPDH [6C5] (mouse; Merck-Millipore, MAB374), 1:1000 contactin-2/TAG-1 (goat; R&D systems, AF4439), 1:1000 CHL1 (goat; R&D systems, AF2147), and 1:1000 N-cadherin [32/N-cadherin] (rabbit; BD, 610920). The following primary antibodies were used for immunostaining: 1:100 NeuN [A60] (mouse; Merck-Millipore, MAB377), 1:200 GFAP (rabbit, Frontier Institute, GFAP-Rb-Af800), 1:200 β-galactosidase (mouse, Promega, Z3781), 1:100 Cux1 (rabbit; Santa Cruz, sc13024), 1:100 Ctip2 (rat; Abcam, ab18465), 1:300 phosphorylated neurofilament [SMI-31] (mouse; Covance, SMI-31R), 1:500 Tbr2 (rabbit; Abcam, ab23345), 1:300 Pax6 (rabbit; BioLegend, PRB-278P), 1:200 phospho-Histone H3 (Ser10) [RR002] (rabbit; Cell Signaling, 9701), and 1:100 Nestin [Rat-401] (mouse; Invitrogen, 14-5843-82). The following secondary antibodies were used for immunostaining: goat anti-rat Alexa Fluor-488, donkey anti-mouse Alexa Fluor-488, donkey anti-rabbit Alexa Fluor-488, donkey anti-mouse Alexa Fluor-594, and donkey anti-rabbit Alexa Fluor-594 (all antibodies are from Life Technologies).
X-Gal staining
Fresh brains were fixed with 2% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) for 30 min–2 h at 4 °C and transferred sequentially to a series of 5, 10, and 30% sucrose in PBS overnight at 4 °C. The brains were embedded in Tissue-Tek OCT compound (Sakura Finetek) and cryosectioned at 30-μm thickness. The sections were mounted on glass slides and washed with solution C (0.01% sodium deoxycholate, 0.02% NP-40, 5 mM EGTA, and 2 mM MgCl2 in PBS) and incubated in solution D (0.5 mg/ml X-gal, 10 mM K3[Fe(CN)6], and 10 mM K4[Fe(CN)6] in solution C) overnight at 37 °C. After fixation with 4% PFA/PBS for 10 min, the sections were subjected to nuclear-fast red staining. Images were acquired using a BX50 microscope (Olympus).
Immunohistochemical analysis
Adult mice were transcardially perfused with 4% PFA/PBS, and the brains were postfixed overnight at 4 °C. For paraffin sections, the fixed brains were dehydrated in 70% ethanol, embedded in paraffin, and sectioned at 4-μm thickness. The sections were subjected to antigen retrieval using an autoclave in 0.01 M citrate buffer at 105 °C for 15 min and incubated with 3% H2O2/MeOH for 30 min. After blocking with 5% BSA/PBS for 30 min, the sections were incubated with anti-NeuN [A60] antibody in 1% BSA/PBS at 4 °C overnight, followed by incubation with EnVision Plus System-HRP Labeled Polymer Anti-Mouse kit (DAKO Cytomation, K4000) for 1 h. The signal was visualized using the Liquid DAB substrate chromogen system (DAKO Cytomation). After rinsing with water, the sections were counterstained with hematoxylin. Images were acquired using a BZ-9000 microscope (Keyence) or a BX50 (Olympus). For quantification of the cortical thickness and the number of NeuN-positive cells, images of 300-μm width cerebral cortex were analyzed using ImageJ software (National Institutes of Health). For bin analysis of the hippocampus, images of 200-μm width × 500-μm length in CA1 and CA3 or 200-μm width × 250-μm length in DG were divided into five equal bins; then, NeuN-positive cell numbers in each bin were counted using ImageJ software (National Institutes of Health). For frozen sections, the brains isolated from E15 embryos and P0 and P6 pups were fixed in 4% PFA/PBS at 4 °C overnight, dehydrated in 30% Sucrose/PBS at 4 °C overnight, embedded in Tissue-Tek OCT compound, and cryosectioned. The 10-μm thick sections were subjected to antigen retrieval in 0.01 M citrate buffer (pH 6.0) at 90 °C for 10 min (for Cux1, Ctip2, Pax6, and Tbr2 staining) or in Histo VT one (NACALAI TESQUE) at 70 °C 10 min (for pHH3 staining), followed by permeabilization with 0.1% Triton X/PBS for 10 min (Pax6 and Tbr2 staining). After blocking with 1% BSA/PBS for 10 min, the sections were incubated with primary antibodies at 4 °C overnight. After washing with PBS, the sections were incubated with 1% BSA/PBS for 10 min, followed by incubation with secondary antibodies and DAPI. The images were acquired using a BZ-9000 microscope (Keyence) or a BX50 (Olympus). Pax6-, Tbr2-, or pHH3-positive cells in 250-μm width CA or 100-μm width DG were analyzed using ImageJ software. Quantitative analyses were conducted blind to genotype. Basically, two sections per animal were analyzed. For β-galactosidase staining, 20-μm thick sagittal sections were rinsed with PBS, permeabilized, and blocked with 0.1% Triton X-100/5% NDS/PBS for 20 min. the sections were incubated with anti-β-galactosidase and anti-NeuN or GFAP antibodies in 0.1% Triton X-100/1% NDS/PBS at 4 °C overnight. After washing with PBS, the sections were incubated with secondary antibodies and DAPI for 1 h. the images were acquired using FV1200 (Olympus).
Measurement of the thickness of the VHC
Coronal sections (4-μm thick) at Bregma −0.35 mm placed on slides were sequentially soaked in xylene (three times for 5 min), 100% ethanol (three times for 3 min), and 95% ethanol (5 min). The sections were then incubated with 0.1% Luxol Fast Blue/95% ethanol at 50 °C overnight. After washing with water, the sections were soaked in 0.05% lithium carbonate for 5 s, dehydrated as above, and counterstained with 0.1% crystal violet (Sigma, C5042). Images were acquired using a BZ-9000 microscope (Keyence) and analyzed using the ImageJ software.
Fluorescent Nissl staining
Briefly, OCT-embedded brains were sectioned in the coronal plane at 50-μm thickness, and slices were transferred to 24-well plates in PBS. The slices were extensively rinsed with PBS and permeabilized with 0.1% Triton X-100/PBS for 20 min. After washing with PBS, the sections were incubated in Neurotrace 530/615 (1:200; Thermo Fisher Scientific; N21482) for 60 min and rinsed with PBS. The images were acquired using a BZ-9000 (Keyence) microscope.
Immunoblot analysis
Isolated mouse brains were homogenized in cold homogenate buffer (50 mM Tris-HCl pH 8.0, 0.32 M sucrose, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride with protease inhibitor cocktail) using a Dounce homogenizer. After the addition of an equal amount of lysis buffer (50 mM Tris-HCl pH 7.5, 250 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 1 mM phenylmethylsulfonyl fluoride with protease inhibitor cocktail), the lysates were incubated on ice for 30 min and centrifuged at 15,000 rpm for 15 min at 4 °C. The supernatants were mixed with the sample buffer, incubated for 30 min at 37 °C, subjected to SDS-PAGE, and blotted onto polyvinylidene fluoride (PVDF) membranes. After blocking with 5% skim milk/TBST (50 mM Tris-HCl pH 8.0, 150 mM NaCl, and 0.05% Tween-20), the membranes were incubated with primary and secondary antibodies in 1% skim milk/TBST. The intensities of the immunoreactive bands were quantified by ImageQuant TL (GE Healthcare).
Hippocampal primary neuron culture
Hippocampi were dissected from E18.5 mouse embryos, treated with 0.15% trypsin (Gibco) and 0.1% Collagenase (Wako) at 30 °C for 30 min, and then triturated gently. In order to measure the lengths of the axons and neurites, 2 × 104 cells (for neurite lengths) or 6 × 104 cells (for contactin-2 intensity) were plated on poly-l-lysine-coated 3.5-cm dishes (for neurite lengths) or coverslips in 3.5-cm dishes (for contactin-2 intensity) and cultured in a neurobasal medium (Gibco, 21103049) supplemented with 2% B27 (Gibco, 17504001) and 0.5 mM glutamine (Sigma-Aldrich) for DIV 3 or DIV 2. To measure neurite length, cells were fixed with 4% PFA/PBS for 30 min, permeabilized with 0.1% Triton-100/PBS for 15 min, blocked with 5% NDS/PBS for 1 h, and incubated with an anti-phosphorylated neurofilament [SMI-31] antibody and rhodamine-conjugated phalloidin (Thermo Fisher Scientific, R415) overnight at 4 °C. The cells were then incubated with a secondary antibody and DAPI for 1 h. Images were acquired using an FV1000 confocal microscope (Olympus). Axon length (phosphorylated neurofilament-positive neurites) and total neurite length (rhodamine-conjugated phalloidin-positive neurites) were measured using the ImageJ software. For contactin-2 staining, cells were fixed with 4% PFA/PBS for 15 min, permeabilized with 0.05% saponin/PBS for 15 min, blocked with 5% NGS/PBS for 1 h, and incubated with an anti-contactin-2 antibody and rhodamine-conjugated phalloidin overnight at 4 °C. Subsequently, the cells were incubated with a secondary antibody and DAPI for 1 h. Images were acquired using an FV1200 confocal microscope (Olympus). To express exogenous GFP and GFP-RAB35, 8 × 104 cells were transfected with pcDNA3.1 GFP or GFP-RAB35 plasmids at DIV 1 using HilyMax (DOJINDO) according to the manufacturer’s instructions and fixed at DIV 3. Quantification of contactin-2 intensity at the somatic plasma membrane was performed using the ImageJ software.
Behavioral analysis
Behavioral tests were performed with the control (Rab35flox/+; Nestin-Cre) and Rab35 cKO (Rab35flox/flox; Nestin-Cre) male littermates at 12–18 weeks of age during the light period of the 12-h light/dark cycle. Mice were handled 5 min per day for 4–5 days prior to a battery of behavioral tests and habituated to the testing room for at least 30 min before each test. Behavioral tests were carried out by an experimenter blinded to the genotype. In the open field test, each mouse was placed in one corner of the open field apparatus, consisting of white PVC (50 × 50 × 40 cm, width × depth × height; O’Hara & Co.) and illuminated at 70 lux, and was allowed to move freely for 15 min. Data were recorded and analyzed using Time OFCR1 software (O’Hara & Co.). The elevated plus maze apparatus (O’Hara & Co.) consisted of two open arms and two closed arms (25 × 5.5 cm each) with 15 cm high transparent walls. The maze was placed 50 cm above the floor and illuminated at 70 lux. Each mouse was placed in the center of the maze facing an open arm and was allowed to move freely in the maze for 10 min. Data were recorded and analyzed using ImageJ EP1 software (O’Hara & Co.). In the rotarod test, mice were placed on a rod (O’Hara & Co.) rotating at 4 rpm. The speed of rotation was accelerated from 4 to 40 rpm over a 300 s period. Mice were trained in three trials per day with a 1.5 h interval between trials for two sequential days. The time at which a mouse fell from the rod was measured. The elevated plus maze apparatus (O’Hara & Co.) consisted of two open arms and two closed arms (25 × 5.5 cm each) with 15 cm high transparent walls. The maze was placed 50 cm above the floor and illuminated at 70 lx. Each mouse was placed in the center of the maze facing an open arm and allowed to move freely for 10 min. Data were recorded and analyzed using the ImageJ EP1 software (O’Hara & Co.). The Y maze apparatus was composed of three opaque arms and illuminated at 70 lx. Each mouse was placed at the end of one arm and allowed to move freely during an 8 min session. The percentage of spontaneous alternations was calculated as follows:
Data were recorded and analyzed using the Time YM1 software (O’Hara & Co.). The Barnes maze apparatus (O’Hara & Co.) is a circular platform (1 m in diameter) with 16 equally spaced holes along the perimeter, elevated 75 cm above the floor. A black escape box was located under one of the holes, which represented the target. The target hole (escape hole) was consistent for each mouse but randomized between mice. Illumination was maintained at 200 lx. Four spatial cues with distinct colors and shapes were hung from the walls of the testing room during the training phase and probe test but not during the habituation phase. During habituation (day 1), each mouse was placed in an opaque cylindrical start chamber at the center of the maze. After 10 s, the mouse was allowed to freely explore the maze for 3 min and then gently guided to the escape box. During the acquisition trials (days 2–5), 10 s after being placed in the start chamber, each mouse was allowed to freely explore the maze for 3 min per trial. Each mouse was given three trials with a 15 min interval between trials for four sequential days. Trials ended when the mouse entered the target hole or when the 3 min had elapsed. Mice that did not enter the escape box were gently guided to the target hole and spent 1 min. During the probe test (day 6), each mouse freely explored the maze without an escape box for 3 min. Data were recorded and analyzed using the Time BCM software (O’Hara & Co.). The probe test used 90 s of 3 min data for analysis.
EdU labeling birthdate analysis
Pregnant mice at E14.5 were intraperitoneally injected with 50 mg/kg body weight of 5-ethynyl-2′-deoxyuridine (EdU; Thermo Fisher Scientific, A10044). The brains of the P12 littermates were isolated and fixed in 4% PFA/PBS at 4 °C overnight, dehydrated in 30% sucrose/PBS at 4 °C overnight, embedded in the OCT compound, and cryosectioned at 20-μm thickness. EdU detection was performed using a Click-it EdU Alexa 594 imaging kit protocol (Thermo Fisher Scientific, C10339) according to the manufacturer’s instructions. Images were acquired by using a BZ-9000 microscope (Keyence). For the quantification, the hippocampus (400-μm width) and cortex (500-μm width) were divided into five or ten equal bins and quantified the number of EdU-positive cells. Basically, two sections per animal were analyzed using ImageJ software.
Hippocampal proteomics
The hippocampi from the control (Rab35flox/+; Nestin-Cre) and Rab35 cKO mice groups at P0 (n = 5 biological replicates per genotype) were lysed in a buffer containing 6 M guanidine hydrochloride, 100 mM HEPES-NaOH (pH 8.0), 10 mM TCEP, and 40 mM chloroacetamide. The lysates were dissolved through heating and sonication, followed by centrifugation at 15,000 rpm for 15 min at 4 °C. Proteins (100 µg each) were purified using methanol/chloroform precipitation and solubilized through the addition of 20 µL of 0.1% RapiGest SF (Waters) in 50 mM triethylammonium bicarbonate. After repeated sonication and vortexing, the proteins were digested with 1 µg of trypsin/Lys-C mix (Promega) for 16 h at 37 °C. The peptide concentrations were determined using the Pierce quantitative colorimetric peptide assay (Thermo Fisher Scientific). Approximately 25 µg of peptides for each sample was labeled with 0.2 mg of TMT10-plex reagents (Thermo Fisher Scientific) for 1 h at 25 °C. After the reaction was quenched with hydroxylamine, all the TMT-labeled samples were pooled, acidified with trifluoroacetic acid (TFA), and fractionated using the Pierce high pH reversed-phase peptide fractionation kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Nine fractions were collected using 5, 10, 12.5, 15, 17.5, 20, 22.5, 25, and 80% acetonitrile (ACN). Each fraction was evaporated in a SpeedVac concentrator and dissolved in 0.1% TFA and 3% ACN.
LC-MS/MS analysis of the resultant peptides (1 µg each) was performed on an EASY-nLC 1200 UHPLC connected to a Q Exactive Plus mass spectrometer through a nanoelectrospray ion source (Thermo Fisher Scientific). The peptides were separated with a linear gradient from 4–20% ACN for min 0–180 and 20–32% ACN for min 180–220, followed by an increase to 80% ACN during min 220–230. The mass spectrometer was operated in a data-dependent acquisition mode with a top 15 MS/MS method. MS1 spectra were measured with a resolution of 70,000, an automatic gain control (AGC) target of 3 × 106, and a mass range of 375–1400 m/z. HCD MS/MS spectra were acquired at a resolution of 35,000, an AGC target of 1 × 105, an isolation window of 0.4 m/z, a maximum injection time of 100 ms, and a normalized collision energy of 34. Dynamic exclusion was set to 30 s. Raw data were directly analyzed against the SwissProt database restricted to M. musculus using Proteome Discoverer version 2.3 (Thermo Fisher Scientific) with Mascot search engine version 2.5 (Matrix Science) for identification and TMT quantification. The search parameters were as follows: (a) trypsin as an enzyme with up to two missed cleavages; (b) precursor mass tolerance of 10 ppm; (c) fragment mass tolerance of 0.02 Da; (d) TMT of lysine and peptide N-terminus and carbamidomethylation of cysteine as fixed modifications; and (e) oxidation of methionine as a variable modification. Peptides were filtered at a false-discovery rate (FDR) of 1% using the percolator node.
To quantify contactin-2 and CHL1, at least three peptides/proteins were measured using PRM, an MS/MS-based targeted quantification method using high-resolution MS. Targeted MS/MS scans were acquired by a time-scheduled inclusion list at a resolution of 70,000, an AGC target of 2 × 105, an isolation window of 4.0 m/z, a maximum injection time of 500 ms, and a normalized collision energy of 27. Time alignment and relative quantification of the transitions were performed using PinPoint version 1.4 (Thermo Fisher Scientific).
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software) and EZR software54. Sample sizes were determined based on previous studies using similar experiments. The experimenters were blinded during the acquisition of imaging data, quantification, and behavior tests. Data, including outliners, were checked for normality using the Shapiro–Wilk test and the equality of variance was checked using F-test. The means between the two groups were compared using a parametric test (two-tailed unpaired Student’s t-test or Welch’s t-test) or a nonparametric test (two-tailed Mann–Whitney U-test). The means between the three groups were compared using a one-way ANOVA. Data from multiple groups were analyzed using two-way repeated-measures ANOVA with Bonferroni multiple comparison tests. In the dot plots, the bars represent mean ± SEM. In the box and whisker plots, the boxes represent the interquartile range (25th to 75th percentile) and the bars in the boxes represent the median. The whiskers cover the range from minimum to maximum. The dots in the graphs show individual data points. Individual information regarding the sample size and statistical tests is described in the figure legends. Statistical significance was set as p < 0.05. All experiments except for the western blot experiment of E13.5 brains and the proteomic analysis were performed at least two times with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data shown in these findings are available from the authors by request. The proteomics data in this study has been deposited in jPOST. The accession numbers are PXD041290 for ProteomeXchange and JPST002112 for jPOST. The original uncropped blots and PCR images can be found in Supplementary Figs. 3, 4. Source data for the graphs can be found in Supplementary Data 2.
References
Zhen, Y. & Stenmark, H. Cellular functions of Rab GTPases at a glance. J Cell Sci. 128, 3171–3176 (2015).
Kouranti, I., Sachse, M., Arouche, N., Goud, B. & Echard, A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol. 16, 1719–1725 (2006).
Chesneau, L. et al. An ARF6/Rab35 GTPase cascade for endocytic recycling and successful cytokinesis. Curr Biol. 22, 147–153 (2012).
Sato, M. et al. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 27, 1183–1196 (2008).
Haley, R., Wang, Y. & Zhou, Z. The small GTPase RAB-35 defines a third pathway that is required for the recognition and degradation of apoptotic cells. PLoS Genet. 14, e1007558 (2018).
Kutscher, L. M., Keil, W. & Shaham, S. RAB-35 and ARF-6 GTPases mediate engulfment and clearance following linker cell-type death. Dev Cell. 47, 222–238.e226 (2018).
Zhang, J., Fonovic, M., Suyama, K., Bogyo, M. & Scott, M. P. Rab35 controls actin bundling by recruiting fascin as an effector protein. Science. 325, 1250–1254 (2009).
Shim, J. et al. Rab35 mediates transport of Cdc42 and Rac1 to the plasma membrane during phagocytosis. Mol Cell Biol. 30, 1421–1433 (2010).
Uytterhoeven, V., Kuenen, S., Kasprowicz, J., Miskiewicz, K. & Verstreken, P. Loss of skywalker reveals synaptic endosomes as sorting stations for synaptic vesicle proteins. Cell. 145, 117–132 (2011).
Brantley, S. E. & Fuller, M. T. Somatic support cells regulate germ cell survival through the Baz/aPKC/Par6 complex. Development. 146, dev169342 (2019).
Hsu, C. et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 189, 223–232 (2010).
Cauvin, C. et al. Rab35 GTPase triggers switch-like recruitment of the Lowe syndrome lipid phosphatase OCRL on newborn endosomes. Curr Biol. 26, 120–128 (2016).
Minowa-Nozawa, A., Nozawa, T., Okamoto-Furuta, K., Kohda, H. & Nakagawa, I. Rab35 GTPase recruits NDP52 to autophagy targets. EMBO J. 36, 2790–2807 (2017).
Patino-Lopez, G. et al. Rab35 and its GAP EPI64C in T cells regulate receptor recycling and immunological synapse formation. J. Biol. Chem. 283, 18323–18330 (2008).
Chevallier, J. et al. Rab35 regulates neurite outgrowth and cell shape. FEBS Lett. 583, 1096–1101 (2009).
Allaire, P. D. et al. Interplay between Rab35 and Arf6 controls cargo recycling to coordinate cell adhesion and migration. J. Cell Sci. 126, 722–731 (2013).
Charrasse, S., Comunale, F., De Rossi, S., Echard, A. & Gauthier-Rouviere, C. Rab35 regulates cadherin-mediated adherens junction formation and myoblast fusion. Mol. Biol. Cell 24, 234–245 (2013).
Kuhns, S. et al. Rab35 controls cilium length, function and membrane composition. EMBO Rep. 20, e47625 (2019).
Kanno, E. et al. Comprehensive screening for novel rab-binding proteins by GST pull-down assay using 60 different mammalian Rabs. Traffic. 11, 491–507 (2010).
Miyamoto, Y., Yamamori, N., Torii, T., Tanoue, A. & Yamauchi, J. Rab35, acting through ACAP2 switching off Arf6, negatively regulates oligodendrocyte differentiation and myelination. Mol. Biol. Cell 25, 1532–1542 (2014).
Sheehan, P., Zhu, M., Beskow, A., Vollmer, C. & Waites, C. L. Activity-dependent degradation of synaptic vesicle proteins requires Rab35 and the ESCRT pathway. J. Neurosci. 36, 8668–8686 (2016).
Villarroel-Campos, D. et al. Rab35 functions in axon elongation are regulated by P53-related protein kinase in a mechanism that involves Rab35 protein degradation and the microtubule-associated protein 1B. J. Neurosci. 36, 7298–7313 (2016).
Chiu, C. C. et al. Increased Rab35 expression is a potential biomarker and implicated in the pathogenesis of Parkinson’s disease. Oncotarget. 7, 54215–54227 (2016).
Vaz-Silva, J. et al. Endolysosomal degradation of Tau and its role in glucocorticoid-driven hippocampal malfunction. EMBO J. 37, e99084 (2018).
Wheeler, D. B., Zoncu, R., Root, D. E., Sabatini, D. M. & Sawyers, C. L. Identification of an oncogenic RAB protein. Science. 350, 211–217 (2015).
Rakic, P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 10, 724–735 (2009).
Hayashi, K., Kubo, K., Kitazawa, A. & Nakajima, K. Cellular dynamics of neuronal migration in the hippocampus. Front. Neurosci. 9, 135 (2015).
Buchsbaum, I. Y. & Cappello, S. Neuronal migration in the CNS during development and disease: insights from in vivo and in vitro models. Development. 146, dev163766 (2019).
Silva, C. G., Peyre, E. & Nguyen, L. Cell migration promotes dynamic cellular interactions to control cerebral cortex morphogenesis. Nat. Rev. Neurosci. 20, 318–329 (2019).
Kawauchi, T. et al. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron 67, 588–602 (2010).
Wu, Q. et al. RAB18, a protein associated with Warburg Micro syndrome, controls neuronal migration in the develo** cerebral cortex. Mol. Brain 9, 19 (2016).
Hor, C. H. H. & Goh, E. L. K. Rab23 regulates radial migration of projection neurons via N-cadherin. Cereb. Cortex 28, 1516–1531 (2018).
Tronche, F. et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99–103 (1999).
Kriegstein, A. & Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184 (2009).
Sun, T. & Hevner, R. F. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat. Rev. Neurosci. 15, 217–232 (2014).
Ayala, R., Shu, T. & Tsai, L. H. Trekking across the brain: the journey of neuronal migration. Cell 128, 29–43 (2007).
Marin, O., Valiente, M., Ge, X. & Tsai, L. H. Guiding neuronal cell migrations. Cold Spring Harb. Perspect. Biol. 2, a001834 (2010).
Belvindrah, R., Nosten-Bertrand, M. & Francis, F. Neuronal migration and its disorders affecting the CA3 region. Front. Cell Neurosci. 8, 63 (2014).
Kawamura, N. et al. Delivery of endosomes to lysosomes via microautophagy in the visceral endoderm of mouse embryos. Nat. Commun. 3, 1071 (2012).
Kawamura, N. et al. Rab7-mediated endocytosis establishes patterning of Wnt activity through inactivation of Dkk antagonism. Cell Rep. 31, 107733 (2020).
Kobayashi, H. & Fukuda, M. Arf6, Rab11 and transferrin receptor define distinct populations of recycling endosomes. Commun. Integr. Biol. 6, e25036 (2013).
Kobayashi, H. & Fukuda, M. Rab35 regulates Arf6 activity through centaurin-beta2 (ACAP2) during neurite outgrowth. J. Cell Sci. 125, 2235–2243 (2012).
Demyanenko, G. P. et al. Close homolog of L1 modulates area-specific neuronal positioning and dendrite orientation in the cerebral cortex. Neuron 44, 423–437 (2004).
Jakovcevski, I. et al. Close homologue of adhesion molecule L1 promotes survival of Purkinje and granule cells and granule cell migration during murine cerebellar development. J. Comp. Neurol. 513, 496–510 (2009).
Katic, J. et al. Interaction of the cell adhesion molecule CHL1 with vitronectin, integrins, and the plasminogen activator inhibitor-2 promotes CHL1-induced neurite outgrowth and neuronal migration. J. Neurosci. 34, 14606–14623 (2014).
Buhusi, M. et al. Close homolog of L1 is an enhancer of integrin-mediated cell migration. J. Biol. Chem. 278, 25024–25031 (2003).
Schlatter, M. C., Buhusi, M., Wright, A. G. & Maness, P. F. CHL1 promotes Sema3A-induced growth cone collapse and neurite elaboration through a motif required for recruitment of ERM proteins to the plasma membrane. J. Neurochem. 104, 731–744 (2008).
Kishimoto, T. et al. Downregulation of L1 perturbs neuronal migration and alters the expression of transcription factors in murine neocortex. J. Neurosci. Res. 91, 42–50 (2013).
Mestres, I., Chuang, J. Z., Calegari, F., Conde, C. & Sung, C. H. SARA regulates neuronal migration during neocortical development through L1 trafficking. Development 143, 3143–3153 (2016).
Denaxa, M., Chan, C. H., Schachner, M., Parnavelas, J. G. & Karagogeos, D. The adhesion molecule TAG-1 mediates the migration of cortical interneurons from the ganglionic eminence along the corticofugal fiber system. Development 128, 4635–4644 (2001).
Kyriakopoulou, K., de Diego, I., Wassef, M. & Karagogeos, D. A combination of chain and neurophilic migration involving the adhesion molecule TAG-1 in the caudal medulla. Development 129, 287–296 (2002).
Malhotra, J. D., Tsiotra, P., Karagogeos, D. & Hortsch, M. Cis-activation of L1-mediated ankyrin recruitment by TAG-1 homophilic cell adhesion. J. Biol. Chem. 273, 33354–33359 (1998).
Sato, T. et al. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature 448, 366–369 (2007).
Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 48, 452–458 (2013).
Acknowledgements
We thank all members of the Sato Laboratory for their kind assistance and helpful discussion. We also thank Drs. Hideaki Miwa, Yuchio Yanagawa (Gunma University), and Akiko Hayashi-Takagi (RIKEN) for their suggestions on mouse behavioral tests; Dr. Masatoshi Nomura (Kurume University) and Drs. Akihiro Harada and Masataka Kunii (Osaka University) for the generation of knockout mouse lines; and Ms. Megumi Kawano (Tokushima University) for her technical support for LC-MS/MS. This work was supported by the JSPS KAKENHI (grant numbers 16J11714, 19J40272, and 23K05678 to I.M.); and the Joint Usage and Joint Research Programs of the Institute of Advanced Medical Sciences, Tokushima University to K.S. This work was also supported by JSPS KAKENHI (17H03669, 19H05711, and 20H00466) and Takeda Science Foundation to K.S.
Author information
Authors and Affiliations
Contributions
K.S., T.H., and I.M. designed the experiments. I.M. characterized the Rab35-knockout mouse lines. K.E., T.K., and H. Koizumi supervised the migration experiments. T.A. and I.I. performed the immunohistochemistry experiments. R.H. and I.M. performed the experiments using primary neurons. T.H., S.T., and H. Kobayashi generated the knockout mouse lines. H. Kosako supervised and performed the mass spectrometry experiments. K.S. and T.H. supervised the project. I.M. and K.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Christian Gonzalez-Billault and Emilie Pacary for their contribution to the peer review of this work. Primary Handling Editors: Edwina McGlinn and George Inglis.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maejima, I., Hara, T., Tsukamoto, S. et al. RAB35 is required for murine hippocampal development and functions by regulating neuronal cell distribution. Commun Biol 6, 440 (2023). https://doi.org/10.1038/s42003-023-04826-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-023-04826-x
- Springer Nature Limited