Abstract
Major depressive disorder (MDD) is a complex condition with unclear pathophysiology. Molecular disruptions within limbic brain regions and the periphery contribute to depression symptomatology and a more complete understanding the diversity of molecular changes that occur in these tissues may guide the development of more efficacious antidepressant treatments. Here, we utilized a mouse chronic social stress model for the study of MDD and performed metabolomic, lipidomic, and proteomic profiling on serum plus several brain regions (ventral hippocampus, nucleus accumbens, and medial prefrontal cortex) of susceptible, resilient, and unstressed control mice. To identify how commonly used tricyclic antidepressants impact the molecular composition in these tissues, we treated stress-exposed mice with imipramine and repeated our multi-OMIC analyses. Proteomic analysis identified three serum proteins reduced in susceptible animals; lipidomic analysis detected differences in lipid species between resilient and susceptible animals in serum and brain; and metabolomic analysis revealed dysfunction of purine metabolism, beta oxidation, and antioxidants, which were differentially associated with stress susceptibility vs resilience by brain region. Antidepressant treatment ameliorated stress-induced behavioral abnormalities and affected key metabolites within outlined networks, most dramatically in the ventral hippocampus. This work presents a resource for chronic social stress-induced, tissue-specific changes in proteins, lipids, and metabolites and illuminates how molecular dysfunctions contribute to individual differences in stress sensitivity.
Similar content being viewed by others
Introduction
Major Depressive Disorder (MDD), a complex, heterogeneous syndrome, is the leading cause of disability worldwide. The symptoms of MDD range from emotional and cognitive impairments as well as systemic dysfunctions. These diverse symptoms suggest the dysregulation of multiple brain regions and peripheral tissues, and there is evidence for brain region-specific disruptions in MDD1. Tricyclic antidepressants (TCAs) initially inhibit serotonin and/or norepinephrine reuptake, but the brain signaling networks affected upon their chronic administration—required for therapeutic efficacy—remain insufficiently understood.
Here we utilize an ethologically-validated mouse model for the study of depression called chronic social defeat stress (CSDS), where mice are exposed chronically to a social stress, which induces a range of MDD-like behavioral and molecular changes in a subset (~ 50%) of animals, referred to as susceptible2,3. These defects are ameliorated by chronic antidepressant treatment2,4,5. The remainder of the stress-exposed population does not display most of these behavioral abnormalities and are referred to as resilient3,4. This divergence in vulnerability to stress is observed within human populations6.
In the present study, we employ metabolomic, lipidomic, and proteomic analyses of the ventral hippocampus (vHipp), nucleus accumbens (NAc), and medial prefrontal cortex (mPFC)—all implicated in MDD—and serum samples from susceptible, resilient, and control (stress naïve) mice in order to comprehensively quantify the changes that occur within these tissues in response to CSDS. We interrogated these brain regions specifically, since each is spatially distinct and contributes uniquely to the limbic system, a circuit of inter-connected brain regions that has been implicated across multiple levels of analysis in the context of depression in both human and animal studies4,7,8. We hypothesized that molecular profiles within these limbic regions would be distinct between resilient and susceptible animals, and that identifying which molecules and pathways are most different would shed light onto the factors responsible for behavioral stress-responses.
Using this approach, we discover that many of the molecules affected by CSDS are involved in the molecular pathways of nucleotide metabolism, fatty acid beta oxidation, and antioxidant function. These pathways are differentially associated with susceptibility vs resilience depending on the brain region involved. We also analyzed the effect of chronic administration of imipramine, a standard tricyclic antidepressant, on these multi-OMIC endpoints. We observe that many of the same pathways are affected by imipramine treatment, further evidence that activity of these pathways contributes to stress responses. Together, this work provides a rich dataset to explore the tissue-specific, molecular mechanisms that differentiate stress resilient and stress susceptible animals, and outlines strongly-affected protein, lipid, and metabolite pathways that present promising targets for antidepressant drug discovery or biomarker efforts.
Methods and materials
Animals
Adult male 7–8 week old C57BL/6 J mice and 6-month old CD1 retired male breeders (CD1 aggressors) were housed at 22–25 °C in a 12-h light/dark cycle and provided food and water ad libitum. All methods were conducted in accordance with the IACUC regulations at Mount Sinai (LA12-00051) and Virginia Commonwealth University (AD10002174). All experiments were approved by the IACUC at these institutions and were performed in accordance with relevant guidelines and regulations.
Chronic social defeat stress and behavioral assays
We utilized an established CSDS protocol as described previously2,3,9. C57BL/6 J mice were exposed for ten consecutive days to a novel aggressive CD1 retired breeder for 10 min and were then separated from the aggressor by a perforated divider to maintain 24 h sensory contact. Each day the test mouse encountered a novel CD1 aggressor mouse. Mice were tested for social interaction (SI) 24 h after the last social defeat by first allowing 2.5 min for the test mouse to explore an arena containing a plexiglass wire mesh cage centered against one wall of the arena (target absent). In the second 2.5 min test, the same test mouse was returned to the arena with a novel CD1 mouse contained in the plexiglass and wire cage (target present). Across all SI tests for a given experiment, the same unfamiliar CD1 (i.e. not used in defeats) target mouse was used to provide consistent social interaction for our test mice. Based on the social interaction ratio, defined as time spent in the ‘interaction zone’ with target present divided by the time spent with target absent, mice were characterized as susceptible (SI ratio < 1) or resilient (SI ratio > 1). The SI ratio of 1 is a commonly used cut-off to discriminate resilient and susceptible animals10,11,12. Control mice were housed identically, yet never came in physical or sensory contact with a CD1 aggressor.
For antidepressant experiments, control, resilient, and susceptible populations were single housed and treated twice-daily with intraperitoneal (IP) injections of saline or imipramine (10 mg/kg) for 14 consecutive days after the SI test5. Treatment-induced changes in MDD-like behaviors were quantified by re-analyzing social interaction behaviors and performing elevated plus maze analysis. For the latter, mice were tested in a standard maze for 10 min, monitored by Ethovision XT as described previously13. Time in the open arms of the plus maze was quantified and expressed as a percent of total time.
Tissue preparation
Twenty-four hours after behavioral testing, vHipp, NAc, and mPFC tissue and serum were collected from consciously decapitated animals, immediately frozen, and stored at − 80 °C. Brains were sectioned in the coronal plane to 1 mm thickness in a brain matrix. Two 16-gauge punches (internal diameter 1.19 mm) were used to microdissect bilateral vHipp, two 14-gauge punches (internal diameter 1.6 mm) were used to isolate NAc bilaterally, and a single 12-gauge tissue punch (internal diameter 2.16 mm) was used to microdissect the mPFC. See Fig. 1A for size and targeting of the tissue punches. Tissue punches were pooled from between 7 and 11 animals depending on the mass of the brain region in order to create a single ~ 20 mg sample—a mass required to run metabolomics, lipidomics, and proteomics in parallel from the same sample. The bilateral vHipp punches of 11 animals were combined to create a single vHipp sample, bilateral NAc punches of eight animals were pooled to create a single NAc sample, and mPFC punches of seven animals were pooled to create a single mPFC sample. The exact number of samples per group is shown in Fig. 1C. The samples were combined in Omni homogenization bead tubes. The sera from two animals were pooled to create 400 µL samples. The aggregation of tissue and serum was performed to equalize SI ratios for samples in each group (resilient SI ratio: ~ 1.4; susceptible SI ratio: ~ 0.8; control SI ratio: ~ 1.35). From these pooled samples, all analyses were performed in parallel. Pooled samples were homogenized in water at 4 °C in an Omni Bead Ruptor 24 (Omni International, Tulsa, OK) and the protein content of each homogenate were determined via a bicinchoninic assay. Aliquots of 100 µg protein, 10 mg tissue weight, and 0.5 mg protein were separated for proteomics, metabolomics, and structural lipidomics analysis, respectively. Aliquots of pooled serum samples were likewise taken for proteomics, metabolomics, structural lipidomics, and mediator lipidomics analysis.
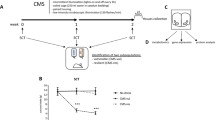
Study overview and metabolomic, lipidomic, and proteomic analysis of serum from resilient and susceptible populations of chronically stressed mice. (A) Graphical illustration of workflow for chronic social defeat stress (CSDS) to differentiate mice into susceptible and resilient populations. All tissues harvested for analysis are displayed. Coronal brain images credit: Allen Institute. (B) Social interaction (SI) data from all mice, tested 24 h after the last CSDS bout. SI ratio is quantified for each mouse, with resilience as an SI ratio > 1 and susceptibility as an SI ratio < 1. Susceptible (n = 37), Resilient (n = 50), and Control (n = 33) (F2,117 = 32.82; ***p < 0.0001; one-way ANOVA followed by Bonferroni post-test). (C) To generate sufficient material for parallel analyses, tissues were pooled by SI ratio to achieve 20 mg/sample for brain tissues and 400 μL/sample for serum. Reported values are the sample numbers for each condition. A single mPFC sample consists of the pooled tissue of seven animals, a single NAc sample consists of the bilateral punches of eight animals, a single vHipp sample consists of the bilateral punches of 11 animals, and a single serum sample is pooled from two animals. (D) From this pooled sample, all processing and analysis occurred in parallel. (E) Heatmap of top 25 affected serum metabolites shows differences in metabolite levels, localized to experimental groups. (F) Pathway analysis of changed metabolites in the serum reveals purine and pyrimidine metabolism, the tricarboxylic acid cycle (TCA cycle), and antioxidant function, among other functions, as significantly affected in the serum of these chronically-stressed mice. Metabolites comprising “Pyrimidine metabolism” network are: Glutamine; Carbamoyl phosphate; Orotidine 5′-phosphate; Uridine; CMP; Cytidine; Deoxycytidine; Deoxyuridine; Thymidine; Thymine; N-Carbamoyl-L-aspartate; Orotate; Uracil; 3-Aminoisobutyrate. “Purine metabolism” network: Xanthine; D-Ribose 5-phosphate; L-Glutamine; 1-(5′-Phosphoribosyl)-5-amino-4-imidazolecarboxamide; AMP; IMP; Adenosine; dAMP; Deoxyadenosine; Deoxyinosine; Xanthosine; Hypoxanthine; Inosine; Guanines; Allantoate; Guanosine; Adenine; Urate; Aminoimidazole ribotide; Urea; Allantoin. “TCA cycle” network: 2-Oxoglutarate; Succinate; Isocitrate; Malate; cis-Aconitate; Citrate; Pyruvate; Fumarate. “Glutathione metabolism” network: Glutathione; NADP + ; Glutathione disulfide; Glycine; L-Glutamate; L-Cysteine; 5-Oxoproline; L-Ornithine; Spermidine. (G) Lipidomic serum analysis reveals total circulating levels of phosphatidic acid (PA) increased in animals resilient to CSDS relative to susceptible animals (F2,58 = 3.80; *p < 0.05; one-way ANOVA followed by Fisher’s LSD comparing resilient and susceptible). (H) Proteomic analysis of serum: a total of 450 proteins were detected, with 17 proteins identified as significantly different from undefeated control animals. Of these 17, three proteins were significantly decreased solely in the susceptible cohort: kallikrein B1 (Klkb1), murinoglobulin-1 (Mug1), and thyroid receptor-interacting protein 11 (Trip11). Images in E, F, and G were generated with MetaboAnalyst 4.0 (https://www.metaboanalyst.ca/).
Targeted and untargeted metabolomic analysis
Metabolomic analyses were performed using gas chromatography–mass spectrometry (GC/MS), reversed-phase liquid chromatography–mass spectrometry (RP-LC/MS), and hydrophilic interaction chromatography–liquid chromatography–tandem mass spectrometry (HILIC-LC/MS/MS)14,15,16,17. Metabolite extraction was achieved using a mixture of isopropanol:acetonitrile:water (3:3:2 v/v/v). Tissue samples were homogenized in an extraction mixture using Fisherbrand Model 120 Sonic Dismembrator. Extract analysis was performed using GC/MS, RP-LC/MS, and HILIC-LC/MS/MS protocols as described14. Quality control was performed using metabolite standards mixtures and pooled samples. The pooled QC sample was obtained by taking an aliquot of the same volume of all samples from the study. Supernatants of tissue and serum extracts were divided in three parts: 75 µL for GC-TOF–MS analysis, 75 µL for RP-LC/MS analysis, and 100 µL for HILIC-LC/MS/MS analysis. Collected raw data were manually inspected, merged, and imputed.
Specifically, metabolomics data were acquired using GC/MS, RP-LC/MS, and HILIC-LC–MS/MS. Recorded mass chromatograms were used for chromatography peak integration. Manual inspection of each mass chromatogram was performed to make sure no failure of separation unit and/or mass spectrometer occurred, and to ensure the acquired data were within the linear range of measurements. Peak areas of the identified metabolites were obtained using vendor’s integration software: ChromaTOF (LECO) and MultiQuant (Sciex). Three datasets containing integrated data for actual batch and all the QCs and pooled samples were merged in a single dataset (i.e. excel sheet) after normalization using external QCs (pooled control human plasma obtained from the UK Biobank, and injected after each 10 samples in a batch) and pooled batch samples was accomplished. Prior normalization missing values imputation was performed with the removal entities having more than 50% of missing or not detected data (N/D or below LOD). Duplicates removal followed with the hierarchy LC–MS/MS > LC–MS > GC–MS based on previously calculated CV for each duplicate detected with using different coupling. Further generalized log (glog) transformation and autoscaling were applied to stabilize the variability of the data. Negative values resulted for peak areas having raw values below 1 (arbitrary unit). Waaijenborg et al. provides additional information on our approaches for fusing metabolomics data from different platforms18.
Statistical analysis was performed with MetaboAnalyst 4.0https://www.metaboanalyst.ca/).
To understand whether the effect of CSDS on these various metabolites was associated with susceptibility or resilience by brain region, we performed untargeted metabolomic profiling in parallel to our targeted analyses. This separate analysis detected many of the same or similar molecules that were identified de novo (Supp. Table 3). Comparing CSDS-exposed mice to undefeated controls, untargeted metabolomics identified 34 metabolites significantly altered in the vHipp, and 11 metabolites significantly altered in the NAc (Supp. Figure 1), again revealing a different metabolic response to stress by brain region. In vHipp the regulation of identified molecules occurred primarily in susceptible animals, whereas in the NAc alterations occurred entirely in resilient animals. Thus, metabolic impact in the NAc is associated with resilience, with metabolic impact in the vHipp associated with susceptibility to chronic stress.
With lipidomic analysis, we observed 36 altered lipid species in the vHipp with multiple species of long chain phosphatidylethanolamine (PE), a major constituent of the myelin sheath, being decreased in susceptible animals (Supp. Table 4A). We identified 54 altered lipid species in the NAc (Supp. Table 4B), and 28 lipid species in mPFC (Supp. Table 4C). With proteomic analysis, we observed 51 altered proteins in the vHipp (Supp. Table 5A), 53 in the NAc (Supp. Table 5B), and 71 in the mPFC (Supp. Table 5C). To interrogate the reliability of our findings, we performed western blot analysis in a separate CSDS cohort for the only protein affected in all three brain regions and the top affected protein in the NAc, RPRD2 (regulation of nuclear pre-mRNA domain containing 2), which plays an important role in gene transcription and was predicted to be elevated in resilient animals relative to susceptible in the NAc. Indeed, we validated this proteomic deduction, which supports the reliability of our analyses (Supp. Figure 2). Further, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis27,28 of the proteome alterations for each brain region (Supp. Table 5D-F).
Imipramine treatment regulates key molecules in the brain and periphery
To investigate the molecular effects of imipramine treatment, we again stratified C57BL/6 J mice into resilient and susceptible populations after CSDS (F2,74 = 18.61; ***p < 0.0001; one-way ANOVA followed by Bonferroni post-test) (Fig. 3A,B) and divided animals into their future treatment groups by equalizing SI ratios. Susceptible, resilient, and control mice were treated twice daily for 14 days with an IP injection of either saline or imipramine (10 mg/kg) (Fig. 3A). Twenty-four hr after the last injection, all mice were examined in a second social interaction test, as well as an elevated plus maze test, to determine the effect of imipramine exposure.
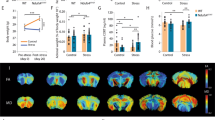
Chronic imipramine treatment improves susceptible behaviors and correlated peripheral molecular profiles. (A) Graphical illustration of workflow for CSDS, treatment schedule, and behavioral testing. Serum, vHipp, NAc, and mPFC were again harvested, pooled by SI ratio, and analyzed as before (see Fig. 1A). (B) SI test 1 data from all mice used to determine susceptible, resilient, and control populations. Animals were pooled into future treatment groups based on equalized SI ratios. Susceptible (n = 33), Resilient (n = 28) and Control (n = 16) (F2,74 = 18.61; ***p < 0.0001; one-way ANOVA followed by Bonferroni post-test). (C) Post-treatment behavioral data including SI and elevated plus maze (EPM). Left: Treatment-induced changes to SI ratios in susceptible population comparing SI test 1 and SI test 2 (n = 14–16; F1,28 = 4.423; * = p < 0.05 by repeated measures two-way ANOVA followed by Bonferroni post-test). Right: EPM test quantifying anxiety-like behaviors in all groups. Resilient animals spend more time exploring the open arms than susceptible animals (F1,56 = 4.19; main effect of group * = p < 0.05 by two-way ANOVA), and treatment significantly improves anxiety-like behaviors (F2,56 = 4.93; main effect of treatment * = p < 0.05 by two-way ANOVA). (D) Metabolomic analysis of serum from imipramine-treated susceptible cohort. Volcano-plot highlights 25 important features. (E) Volcano plot of the 11 affected lipids. (F) Volcano plot reveals five affected proteins. n = 8 samples per group for panels D-F. Volcano plots in D-F were generated with MetaboAnalyst 4.0 (https://www.metaboanalyst.ca/).
As expected5, chronic imipramine administration improved social interaction deficits seen in susceptible mice. We compared the SI ratio from test 1 (before treatment) and SI test 2 (after treatment), and while saline treatment did not improve sociability, there was a significant improvement in SI ratio in imipramine-treated susceptible mice (n = 14–16; F1,28 = 4.423; * = p < 0.05 by repeated measures two-way ANOVA followed by Bonferroni post-test) (Fig. 3C).
We next performed the elevated plus maze assay. We observed a significant difference in time spent on the open arm between susceptible and resilient groups, with the former group spending less time in the open arm, indicating that group phenotypes persisted throughout the treatment window (F1,56 = 4.19; main effect of group * = p < 0.05 by two-way ANOVA). We also observed a significant treatment effect in both susceptible and resilient groups with imipramine treatment increasing the amount of time spent exploring the open arm (F2,56 = 4.93; main effect of treatment * = p < 0.05 by two-way ANOVA). (Fig. 3C). Collectively, these data indicate that imipramine improves behavioral abnormalities induced by CSDS. To understand the molecular pathways affected by imipramine exposure in chronically-stressed mice, we performed multi-OMIC profiling on pooled samples of serum and of vHipp, NAc, and mPFC of all mice.
In the serum of susceptible animals treated with saline or imipramine, metabolomic analysis identified 25 significantly affected metabolites (Fig. 3D, identities in Supp. Table 6A). Notably, imipramine treatment reduces the levels of many metabolites, with many involved in previously identified functions including the antioxidant precursor gamma-glutathione-cysteine and the coenzyme flavin adenine dinucleotide (FAD) which participates in many metabolic functions including the TCA cycle.
With lipidomic analysis, we observed that imipramine treatment significantly affected the serum concentrations of 11 lipids (Fig. 3E, identities in Supp. Table 6B): sphingomyelin and lysophosphatidylserine species were decreased, and there was an increase in a PA species, which is consistent with the earlier-observed increase in PA species specifically in resilient animals.
Lastly, with proteomic analysis, we identified five significantly affected proteins (Fig. 3F, identities in Supp. Table 6F). Kallikrein 1-related peptidase b5 (Klk1b5) and Kallikrein 1-related peptidase b24 (Klk1b24) were each increased by imipramine treatment. This is consistent with our earlier observation of decreases in Klkb1 in the serum of susceptible animals (Fig. 1H), suggesting that imipramine may act to increase Klkb1-related functions. This observation highlights the potentially important role for peripheral protease activity in contributing to stress responses.
We then performed metabolomic analysis in each brain region. In the vHipp, as in serum, we observed primarily a down-regulation of key metabolites (including AICAR involved in de novo purine synthesis, the antioxidant glutathione disulfide, S1P, and acetylphosphate), with nine metabolites significantly affected (Fig. 4A, list in Supp. Table 7A). A heatmap of the top 25 affected metabolites clusters by treatment, revealing how imipramine treatment affects the vHipp and acts to diminish the levels of metabolites that were previously observed to be elevated in the vHipp of susceptible animals (Supp. Figure 3). In the NAc, four metabolites are affected (Fig. 4B, list in Supp. Table 7B). Lastly, only one metabolite was significantly affected in the mPFC (Fig. 4C, list in Supp. Table 7C). These data mirror the observations of Fig. 2, in that the brain areas that were most affected metabolically by stress are also most affected by imipramine treatment.
Imipramine-induced molecular adaptations by brain region within susceptible animals. (A) Volcano plot of nine changing metabolites identified in the vHipp. n = 3 samples per group. (B) Volcano plot of four changing metabolites identified in the NAc. n = 5 samples per group. (C) Volcano plot of one changing metabolite identified in the mPFC. n = 5 samples per group. Identities and statistics for all metabolites can be found in Supplemental Table 7A-C. Volcano plots in A-C were generated with MetaboAnalyst 4.0 (https://www.metaboanalyst.ca/).
Metabolic network map**
We utilized the metabolomics data in serum and brain to construct the net effect of CSDS and imipramine treatment on metabolic pathways. In the vHipp, we observed consistent changes in many of the metabolites within the de novo purine synthesis pathway with resilient animals showing decreases and susceptible animals showing increases in these metabolites (Fig. 5A). Many purine products are decreased in susceptible animals. ADP, a precursor of ATP, is significantly elevated in susceptible and decreased in resilient animals. A key product of ATP, s-adenosyl methionine (SAM-e) is diminished in the vHipp of resilient animals. SAM-e is a major methyl donor used for protein and lipid methylation, and differences in its bioavailability within the vHipp could have swee** consequences on cellular functions.
Ventral hippocampus metabolic networks differentially affected in susceptibility vs resilience. Red and blue arrows indicate regulation of metabolite levels observed across our datasets in resilient and susceptible animals, respectively. The test tube icon represents an alteration observed in serum samples, as these molecules are excreted in urine. The hyperactive pathway through these networks experienced by susceptible animals is highlighted in blue. Metabolite relationships were derived from KEGG database and the IUBMB-Sigma-Nicholson Metabolic Pathways Chart. Solid black arrows represent direct metabolic reactions. Yellow boxes represent basic cellular functions affected by key metabolites. Molecules that survived FDR correction are in bold. Molecules affected by imipramine treatment are italicized. PRPP, phosphoribosyl pyrophosphate; GAR, glycinamide ribonucleotide; FGAR, 5′-Phosphoribosyl-N-formylglycineamide; AICAR, 5-Aminoimidazole-4-carboxamide ribonucleotide; IMP, inosine monophosphate; IDP, inosine diphosphate; XMP, xanthosine monophosphate; sAMP, succinyl adenosine monophosphate; AMP, adenosine monophosphate; ADP, adenosine diphosphate; ATP, adenosine triphosphate.
In the vHipp, beyond the purine network, susceptible and resilient animals show opposing effects in fatty acid beta oxidation pathways. Susceptible animals show hyperfunction of the fatty acid beta oxidation cycle (Fig. 5B). Beta oxidation is the process through which fatty acids are internalized in the mitochondria via carnitine modification and consumed to yield acetyl-CoA29. These susceptible-specific effects in glycolysis and beta oxidation converge in the accumulation of acetyl-CoA, further implicating hyper-energetics in the vHipp of susceptible animals. Also, acetyl-CoA is a source for protein acetylation, including histones30. This potential regulation of histone acetylation levels is consistent with a literature in both mice and rats describing changes in hippocampal histone acetylation in response to stress and antidepressant exposure5,31,32,33.
We were unable to resolve significant differences in ATP or acetyl-CoA themselves, likely due to the fact that these metabolites are consumed rapidly and utilized for numerous cellular functions. However, resolving the full metabolic network, including precursor and product molecules, serves to reveal metabolic demands. Taken together, these analyses demonstrate that the brain-region specific cellular functions of energy production and the global post-translational modifications of methylation and acetylation are likely hyperactive in the vHipp of susceptible animals. This illuminates that individual differences in these cellular functions by brain-region may underlie the divergence in stress responses.