Abstract
Supplemental energy and protein during calf-hood (2–30 wk) in dairy bulls hastened puberty (~1 mo), upregulated steroid biosynthesis, concentrations of reproductive hormones and Sertoli cell maturation, with larger testes and greater sperm production (~25%) in mature bulls. The objective was to evaluate effects of feeding high (20.0% crude protein [CP], 67.9% total digestible nutrients [TDN]), control/medium (17.0% CP, 66.0% TDN) and low (12.2% CP, 62.9% TDN) diets from 2 to 30 wk on post-pubertal testes of Holstein bulls. Based on RNA sequencing, 497 and 2961 genes were differentially expressed (P < 0.1) in high- vs low- and high- vs medium-diet groups, respectively. According to KEGG analysis, oxidative phosphorylation and ribosome pathways were upregulated in high- vs medium- and low-diet groups, with majority of upregulated genes encoding for essential subunits of complex I, III, IV and V of OXYPHOS pathway. In addition, mitochondrial translation, mitotic nuclear division and cell division were enriched in high- vs medium-diet groups. Consistent with these results, a greater percentage of sperm from high-diet bulls were progressively motile and had normal mitochondrial function compared to medium-diet sperm (P < 0.1). Thus, enhanced early life nutrition upregulated mitochondrial function in testes and sperm of post-pubertal Holstein bulls.
Similar content being viewed by others
Introduction
Enhanced early-life nutrition hastens puberty, with ~25% increases in testis size and sperm production in dairy and beef bulls1,2. The underlying endocrine basis is profound increases in luteinizing hormone (LH) pulse frequency and insulin like growth factor (IGF-I) concentrations in blood3,4. In a cohort-based study in Sweden, low nutrition during the pre-pubertal period in men reduced susceptibility to heart disease in their offspring5, consistent with developmental programming in male germ cells that extends well into early post-natal life6. Therefore, nutritional modulation during early life affects testicular development, with apparent epigenetic effects on post-pubertal sperm function.
We reported increased cholesterol/steroid biosynthesis and Sertoli cell maturation in testicular tissues of high-diet bulls at 16 and 24 wk, respectively7. Although a wide range in pre-pubertal diets had no significant effects on routine analyses of sperm morphology and function8, molecular-level analyses should be done, to ensure no deleterious effects are transmitted to the next generation. Our objective was to determine effects of pre-pubertal dietary modulations on post-pubertal testes of Holstein bulls.
Results
mRNA profile of testes
Approximately 340 × 106 reads were obtained from the 24 libraries sequenced. On average, 14,357,890 (SD = 2,267,076) reads were obtained per library and 13,565,597 (SD = 2,148,261) mapped to the Ensemble gene annotation database. On average 13,710,936 (SD = 3,098,630), 13,120,341 (SD = 1,219,498) and 13,865,513 (SD = 1,929,109) reads were mapped from HD, MD and LD groups respectively. A total of 18,288 genes were detected in testicular tissue and their Ensembl IDs are provided (Supplementary Dataset 1). On average, the most abundant gene in the testicular transcriptome was cytochrome c oxidase subunit I (COX1), a mitochondrial gene (ENSBTAG00000043561), representing ~2.9% of total reads. Other abundant genes included cytochrome c oxidase subunit 3 (COX3, ENSBTAG00000043560), ATP synthase F0 subunit 6 (ATP6, ENSBTAG00000043584) and cytochrome b (CYB, ENSBTAG00000043550), all mitochondrial genes.
Differentially expressed genes
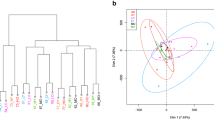
In general, of 18288 genes expressed, 2960 were differentially expressed (P < 0.1) between HD and MD groups compared to 496 in HD versus LD groups (Supplementary Datasets 2 and 3). A scatterplot denoting transcriptomic differences across diet groups is provided (Supplementary File 1). To visualise extent of differential expression and relationship between mean gene expression levels and log2fold changes, MA plots were generated (Fig. 1). Of the 496 DEGs in the HD versus LD analysis, 446 genes were upregulated in the HD group (genes with higher log2fold change were: COX5B, COX6B1, ATP5J, ATP5G1, MRPL11 and HSD17B1). When comparing HD and MD groups, of the 2960 DEGs, 1828 genes were upregulated in the HD group (genes with higher log2fold change: COX2, COX3, COX5B, HSD17 and CCNA1). The log2fold changes varied from ± 0.21 to ± 1.17 among all DEGs. No genes were differentially expressed between MD and LD groups. Therefore, all further analyses were done using two datasets, HD vs LD and HD vs MD groups.
Gene Ontology term enrichment
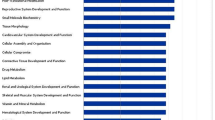
To systematically examine enriched genes associated with differential pre-pubertal diets on post-pubertal testes, gene ontology (GO) analysis was done using the functional annotation and clustering option in DAVID. “Mitochondrial translation initiation” and “elongation” were the major biological functions enriched in HD vs LD and HD vs MD groups. In the HD vs MD group comparison, “mitotic nuclear division”, “cytoplasmic translation” and “ATP synthesis coupled protein transport” were also enriched. Complete lists of GO terms enriched in HD vs LD and HD vs MD datasets are provided in Tables 1 and 2, respectively.
Over-represented KEGG pathways
Eight pathways were associated with HD vs MD groups and six pathways in HD vs LD groups when considering genes (P < 0.1, Tables 3 and 4). “Oxidative phosphorylation” and “ribosome pathways” were major pathways enriched in both datasets.
In addition to above, oxidative phosphorylation and ribosome pathways were enriched in the upregulated genes from both datasets (HD vs MD and HD vs LD, Supplementary Files 2 and 3). However, nine downregulated pathways were identified in the HD vs MD dataset (Supplementary File 4). Major pathways identified were focal adhesion, regulation of actin cytoskeleton and axon guidance.
PCR validation of differentially expressed genes
Five genes were selected from the list of differentially expressed mRNA for validation (Table 5). We were able to successfully validate four (CRHR1, TJP2, ATP5G1 and ATP5J) of the five genes selected (P < 0.1, Fig. 2).
Sperm progressive motility and mitochondrial membrane potential (MMP)
Sperm of HD bulls had greater (P < 0.1) post-thaw progressive motility than sperm from MD or LD bulls (22.3 ± 3.0, 8.7 ± 2.9 and 10.8 ± 3.0 respectively, Fig. 3). There were also differences across dietary treatments in percentages of Rhl23-/PI- (P < 0.1) and Rhl23-/PI + sperm (P < 0.05; Table 6). Percentage of Rhl23−/PI− sperm (live sperm loosing mitochondrial function) was higher in MD vs HD groups (15.1 ± 2.6 vs 8.6 ± 0.9). However, there was no significant difference among groups for percentage of Rhl23 + /PI- sperm (live sperm with normal mitochondrial function). The percentage of Rhl23-/PI + sperm (dead or necrotic sperm) in the MD was 66.2 ± 3.5 and was lower (P < 0.05) than both LD and HD groups (78.6 ± 2.2 and 78.7 ± 2.2, respectively). A representative image of flow cytometry explaining sperm populations is provided (Fig. 4).
Representative images of flow cytometry using sperm stained with Rhodamine 124 and PI to evaluate mitochondrial membrane potential (MMP). (a) Represents the gating to establish the population (P1); (b) histogram for SYBR14; (c) histogram for PI; (d) shows the four population of cells- Lower right quadrant (Q4): Rh123 + /PI−, live sperm with normal mitochondrial function, Lower left quadrant (Q3): Rh123−/PI−, live sperm losing mitochondrial function, Upper left quadrant (Q1): Rh123−/PI+, dead/necrotic sperm.
Discussion
To provide a more comprehensive assessment of post-pubertal effects of pre-pubertal feeding, RNA-sequencing was done on testicular tissues collected at 72 wk of age. To our understanding, this is the first study to evaluate effects of differential feeding during early pre-pubertal period on the post-pubertal bovine transcriptome using a next-generation sequencing technique. Bulls on the HD (fed supplemental protein and energy) had greater oxidative phosphorylation and mitochondrial protein synthesis within their testes compared to LD and MD bulls. Furthermore, sperm from HD bulls had greater progressive motility and a greater percentage had normal mitochondrial function compared to sperm from MD bulls.
Oxidative phosphorylation (OXYPHOS) occurs in mitochondria, generating nearly 80% of cellular energy. OXYPHOS, the functional unit of mitochondria, connects the electron transport chain (ETC) to cell respiration and ATP synthesis. Enzyme complexes constituting OXYPHOS are comprised of mitochondrial and nuclear encoded polypeptides9. Elevated mitochondrial and cytoplasmic translation in HD vs MD were indicative of increased protein availability for OXYPHOS formation. A similar study in Holstein bulls also identified increased OXYPHOS in adipose tissue along with enhanced testes development, cholesterol and androgen biosynthesis in the testes of pre-pubertal bulls fed high vs low planes of nutrition10,11,12.
The integral part of OXYPHOS, the ETC, is composed of five multi-protein complexes and two electron carriers (ubiquinone and cytochrome) transporting electrons to oxygen, creating a proton motive force. The resulting membrane potential enables energy production when these protons flow back via ATP synthase13. Proton transfer at sites other than ATP synthase reduces mitochondrial efficiency, but is critical for ROS production. Testicular mitochondria consume less oxygen than other tissues to maintain the same electric potential and have higher phosphorylation efficiency with age14. In mature testes, spermatocytes and spermatids use OXYPHOS, due to limited glucose availability in seminiferous tubule fluid, whereas Sertoli cells, spermatocytes and mature sperm have high glycolytic activity15,16. The majority of upregulated genes in HD (vs MD and LD) were significantly enriched for OXYPHOS.
In sperm, mitochondrial activity has been associated with sperm motility17, viability18, capacitation19 and fertilization potential20. Within germ cells, mitochondrial ATP synthesis was reported to regulate the apoptotic pathway21. Higher MMP has been associated with increased sperm motility and production, as sperm require ATP17. In addition, MMP is correlated with activity of the ETC and ATP synthase (components of the OXYPHOS). Increased mitochondrial activity beyond its threshold could induce oxidative stress, which could alter DNA. The majority of genes involved in the ETC had high expression in HD bulls compared to MD and LD bulls. In addition, major mitochondrial functions, including ATP synthesis coupled protein transport, hydrogen ion transmembrane transport, mitochondrial electron transport and oxidation reduction processes were altered in HD vs MD bulls.
A positive association between mitochondrial function and motility has been reported in bovine sperm22. In our study, sperm from HD bulls had greater progressive motility (PM) compared to sperm from MD and LD bulls; the association between motility and mitochondrial function, reflected our gene expression study. Furthermore, positive associations between OXYPHOS, ETC and sperm motility have been reported18,23,24. We also evaluated MMP, for which Rhodamin123, a mitochondria-specific fluorescent dye in combination with PI, was used43 and with Bowtie v.1.1.244 used as an internal aligner. Map** rate was uniformly good, with >93% of reads successfully mapped to the genome.
Detection of differentially expressed genes
Counts of reads map** to genes were obtained using featureCounts software from Subread package45. Count data were loaded into R, with normalization and variance stabilizing transformation46 applied using DESeq2 package. Normalized and variance stabilized data were used for quality control and sample clustering analyses. Euclidean sample distances were calculated in R and sample clustering was done using hclust47 function in R. Sample to sample distances were visualized as heatmaps. Principal components plots were made with PCAplot function in DESeq2 package48.
Genes differentially expressed between diets were detected using Wald tests on normalized count data. Experimental conditions were compared with one-way ANOVA; thereafter, results for specific contrasts (HD vs LD, HD vs MD and MD vs LD) were extracted. Correction procedures for multiple comparisons were done with Benjamini-Hochberg correction implemented in R. Genes with adjusted P-values < 0.1 were considered differentially expressed.
Functional analysis
The Ensembl identities of DEGs from HD vs LD and HD vs MD analyses were uploaded to Database for Annotation, Visualization and Integrated Discovery (DAVID) to enrich for Gene Ontology (GO) Biological process and cellular components49. For most analyses, we used the functional annotation and clustering option and reported all GO terms that were P < 0.05 and molecule number ≥2.
Quantitative real-time polymerase chase reaction (RT-PCR)
RT-PCR was performed as described previously7. In brief, reverse transcription and subsequently PCR with SYBR Green (Fast SYBR® Green Master Mix; Applied Biosystems, Foster City, CA, USA) was done to validate mRNA expression of five differentially expressed genes: TJP2, COX5B, ATP5J, ATP5G1 and CRHR1. The National Center for Biotechnology and Information (NCBI) was used to design PCR primer sequences (Table 5), with primers purchased from Thermo Fisher Scientific (Waltham, MA, USA). RNase-free DNase I (Thermo Fisher Scientific) was added to total RNA (1 µg). After incubation at RT for 15 min at RT, 25 mM EDTA was added and the mixture hated to 65 °C for 10 min to cause denaturation. Then, High Capacity cDNA Reverse Transcription Kit and random hexamers (Applied Biosystems) were used for reverse transcription. For all primers, cDNA was diluted 1:10 and was efficiently used for PCR. All amplification reactions were done in 96 well plates (Applied Biosystems), as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 59 °C for 1 min and finally dissociation (55–95 °C). Every reaction well contained: Fast SYBR green Master mix- 7.5 μl, forward primer (20 pmol/μl)- 0.5 μl, reverse primer (20 pmol/μl)- 0.5 μl, cDNA template- 0.5 μl and water-1 μl. All reactions were done in duplicate, with inclusion of appropriate controls (non-RT and no-cDNA). Purity was confirmed by melt curve analysis. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH;50) was used as a housekee** gene to normalize Cq (mean threshold cycle) and (∆) Cq values were expressed as 1/ ∆Cq for ease of understanding interpretation of gene expression.
Evaluation of sperm progressive motility
Sperm collected from a subset of three bulls/treatment at two time points (55–59 and 69–71 wk) were analyzed; this selection of bulls and time points was based on availability of cryopreserved semen samples. Sperm progressive motility was evaluated using computer assisted sperm analysis (CASA), as described51. In brief, a small aliquot of semen (4 μL) was loaded into a prewarmed chamber slide (20 μm deep; Leja Products, Nieuw-109, Vennep, the Netherlands) and immediately analyzed under 400X magnification for progressive motility. Seven microscopic fields per sample were analyzed and the frame rate was 60/s.
Evaluation of sperm mitochondrial membrane potential (MMP)
Sperm samples used for motility analysis were also assessed for mitochondrial membrane potential with a combination of Rhodamine 123 and Propidium Iodide (PI). Rh123/PI dual fluorescent staining and flow cytometry were done as described25. In brief, frozen-thawed semen was washed twice in PBS (700 x G for 5 min) and sperm concentration adjusted to 5 × 106/mL. Then, Rh123 (Molecular Probes, Eugene, OR, USA) was added to the sperm suspension at a final concentration of 5 µg/mL and incubated for 5 min at 37 °C in the dark. To remove non-specific binding, sperm were washed in PBS (700 x G for 10 min), followed by addition of propidium iodide (PI, Invitrogen, Canada, Burlington, ON, Canada) at a final concentration of 5 µg/mL and incubated for 5 min in the dark. The PI staining was performed for concurrent evaluation of both viability and mitochondrial function. Flow cytometry analysis (FACScan, Beckon Dickinson, San Jose, CA, USA) was done with an argon excitation laser (488 nm) and >10 × 103 events recorded for each sample.
Statistical analyses
Data for RT-qPCR, motility and mitochondrial membrane potential were analysed using R software (v 3.4.3;47). Depending on number of groups involved, ANOVA or independent Student’s t-tests were used to detect differences, with P < 0.1 defined as significant.
Data availability
All data generated or analysed during this study were deposited in the publicly available NCBIs Gene Expression Omnibus Database GSE137690 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE137690.
References
Barth, A. D., Brito, L. F. C. & Kastelic, J. P. The effect of nutrition on sexual development of bulls. Theriogenology 70, 485–494 (2008).
Brito, L. F. C. et al. Circulating metabolic hormones during the peripubertal period and their association with testicular development in bulls. Reprod. Domest. Anim. 42, 502–508 (2007).
Brito, L. F. C. et al. Effect of improved nutrition during calfhood on serum metabolic hormones, gonadotropins, and testosterone concentrations, and on testicular development in bulls. Domest. Anim. Endocrinol. 33, 460–469 (2007).
Dance, A., Thundathil, J., Wilde, R., Blondin, P. & Kastelic, J. Enhanced early-life nutrition promotes hormone production and reproductive development in Holstein bulls. J. Dairy Sci. 98, 987–998 (2015).
Kaati, G., Bygren, L. O. & Edvinsson, S. Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents’ slow growth period. Eur. J. Hum. Genet. 10, 682–688 (2002).
Oakes, C. C., La Salle, S., Smiraglia, D. J., Robaire, B. & Trasler, J. M. Developmental acquisition of genome-wide DNA methylation occurs prior to meiosis in male germ cells. Dev. Biol. 307, 368–379 (2007).
Johnson, C., Dance, A., Kovalchuk, I., Kastelic, J. & Thundathil, J. Enhanced early-life nutrition upregulates cholesterol biosynthetic gene expression and Sertoli cell maturation in testes of pre-pubertal Holstein bulls. Sci. Rep. 9, 6448 (2019).
Dance, A., Thundathil, J., Blondin, P. & Kastelic, J. Enhanced early-life nutrition of Holstein bulls increases sperm production potential without decreasing postpubertal semen quality. Theriogenology 86, 687–694.e2 (2016).
Taanman, J. W. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta - Bioenerg. 1410, 103–123 (1999).
English, A. M. The molecular control of the hypothalamic-pituitary-testicular axis in the bull. (University of Limerick, 2018).
English, A. M. et al. Role of early life nutrition on regulating the hypothalamic-anterior pituitary-testicular axis of the bull. Reproduction 156, 283–297 (2018).
English, A. M. et al. Effect of Early Calf-Hood Nutrition on the Transcriptional Regulation of the Hypothalamic-Pituitary- Testicular axis in Holstein-Friesian Bull Calves. Sci. Rep. 8, 1–10 (2018).
Bottje, W. G. & Carstens, G. E. Variation in Metabolism: Biological Efficiency of Energy Production and Utilization That Affects Feed Efficiency. Feed Efficiency in the Beef Industry (2012).
Ramalho-Santos, J. et al. Mitochondrial functionality in reproduction: from gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update 15, 553–572 (2009).
Robinson, R. & Fritz, I. B. Metabolism of Glucose by Sertoli Cells in Culture. Biol. Reprod. 24, 1032–1041 (1981).
Meinhardt, A. et al. A switch in the cellular localization of macrophage migration inhibitory factor in the rat testis after ethane dimethane sulfonate treatment. J. Cell Sci. 112, 1337–1344 (1999).
Deffosez, A., Marchetti, C., Obert, G., Marchetti, P. & Formstecher, P. Study of mitochondrial membrane potential, reactive oxygen species, DNA fragmentation and cell viability by flow cytometry in human sperm. Hum. Reprod. 17, 1257–1265 (2002).
Ruiz-Pesini, E. et al. Seminal quality correlates with mitochondrial functionality. Clin. Chim. Acta. 300, 97–105 (2000).
Stendardi, A. et al. Evaluation of mitochondrial respiratory efficiency during in vitro capacitation of human spermatozoa. Int. J. Androl. 34, 247–255 (2011).
Kasai, T. et al. Relationship between sperm mitochondrial membrane potential, sperm motility, and fertility potential. Asian J. Androl. 4 (2002).
Erkkila, K. et al. Regulation of human male germ cell death by modulators of ATP production. Am. J. Physiol. Metab. 290, E1145–E1154 (2006).
Garner, D. L., Thomas, C. A., Joerg, H. W., DeJarnette, J. M. & Marshall, C. E. Fluorometric Assessments of Mitochondrial Function and Viability in Cryopreserved Bovine Spermatozoa1. Biol. Reprod. 57, 1401–1406 (1997).
John, J. C. S. T., Jokhi, R. P. & Barratt, C. L. R. The impact of mitochondrial genetics on male infertility. Int. J. Androl. 28, 65–73 (2005).
Ruiz-Pesini, E. et al. Correlation of sperm motility with mitochondrial enzymatic activities. Clin. Chem. 44, 1616 LP–1620 (1998).
Zou, T., Liu, X., Ding, S. & **ng, J. Evaluation of sperm mitochondrial function using rh123/PI dual fluorescent staining in asthenospermia and oligoasthenozoospermia. J. Biomed. Res. 24, 404–410 (2010).
Marchi, S. et al. Mitochondria-ros crosstalk in the control of cell death and aging. J. Signal Transduct. 2012, 329635 (2012).
Wang, X. et al. Alterations in mitochondria membrane potential and oxidative stress in infertile men: a prospective observational study. Fertil. Steril. 80, 844–850 (2003).
Rajender, S., Rahul, P. & Mahdi, A. A. Mitochondria, spermatogenesis and male infertility. Mitochondrion 10, 419–428 (2010).
Thomson, E., Ferreira-Cerca, S. & Hurt, E. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 126, 4815–4821 (2013).
Hales, D. B. et al. Mitochondrial Function in Leydig Cell Steroidogenesis. Ann. N. Y. Acad. Sci. 1061, 120–134 (2005).
Farkash, Y., Timberg, R. & Orly, J. Preparation of antiserum to rat cytochrome P-450 cholesterol side chain cleavage, and its use for ultrastructural localization of the immunoreactive enzyme by protein A-gold technique. Endocrinology 118, 1353–1365 (1986).
Stocco, D. M. StAR Protein and the Regulation of Steroid Hormone Biosynthesis. Annu. Rev. Physiol. 63, 193–213 (2001).
Matilainen, O., Quirós, P. M. & Auwerx, J. Mitochondria and Epigenetics – Crosstalk in Homeostasis and Stress. Trends Cell Biol. 27, 453–463 (2017).
Minocherhomji, S., Tollefsbol, T. O. & Singh, K. K. Mitochondrial regulation of epigenetics and its role in human diseases. Epigenetics 7, 326–334 (2012).
Holliday, R. & Pugh, J. E. DNA modification mechanisms and gene activity during development. Science 187, 226–232 (1975).
Turner, M. B. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. CMLS 54 (1998).
Wallace, C. D. & Fan, W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion 10 (2010).
Gonzalez-Sanchez, E., Marin, J. J. G. & Perez, M. J. The Expression of Genes Involved in Hepatocellular Carcinoma Chemoresistance Is Affected by Mitochondrial Genome Depletion. Mol. Pharm. 11, 1856–1868 (2014).
Champroux, A., Cocquet, J., Henry-Berger, J., Drevet, J. R. & Kocer, A. A Decade of Exploring the Mammalian Sperm Epigenome: Paternal Epigenetic and Transgenerational Inheritance. Front. cell Dev. Biol. 6, 50 (2018).
Donkin, I. & Barres, R. Sperm epigenetics and influence of environmental factors. Mol. Metab. 14, 1–11 (2018).
Schagdarsurengin, U. & Steger, K. Epigenetics in male reproduction: effect of paternal diet on sperm quality and offspring health. Nat. Rev. Urol. 13, 584–595 (2016).
Andrews, S. FastQC: a quality control tool for high throughput sequence data. (2010).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Durbin, B. P., Hardin, J. S., Hawkins, D. M. & Rocke, D. M. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics 18, S105–S110 (2002).
Team, R. D. C. R: A Language and Environment for Statistical Computing. (2008).
Varet, H., Brillet-Gueguen, L., Coppee, J. Y. & Dillies, M. A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS One 11, e0157022 (2016).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 (2009).
Klein, C., Scoggin, K. E., Ealy, A. D. & Troedsson, M. H. T. Transcriptional Profiling of Equine Endometrium During the Time of Maternal Recognition of Pregnancy. Biol. Reprod. 83, 102–113 (2010).
Krishnakumar, S., Whiteside, D., Dance, A. & Elkin, B. T. J. Effect of Chilling Duration on Post-Thaw Characteristics of Sperm from the North American bison (Bison bison). Reprod Domest Anim 48, 636–42 (2013).
Author information
Authors and Affiliations
Contributions
C.J. wrote the entire manuscript and conducted the experiments with I.K. The experiment was conceived and designed by C.J. and A.D. Data analysis was performed by I.K. and interpreted by C.J., A.D., J.K. and J.T. designed the animal work component. All co-authors reviewed and agreed for submission of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Johnson, C., Dance, A., Kovalchuk, I. et al. Enhanced pre-pubertal nutrition upregulates mitochondrial function in testes and sperm of post-pubertal Holstein bulls. Sci Rep 10, 2235 (2020). https://doi.org/10.1038/s41598-020-59067-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-59067-3
- Springer Nature Limited
This article is cited by
-
Prepubertal nutritional modulation in the bull and its impact on sperm DNA methylation
Cell and Tissue Research (2022)