Abstract
3D microstructural datasets are commonly used to define the geometrical domains used in finite element modelling. This has proven a useful tool for understanding how complex material systems behave under applied stresses, temperatures and chemical conditions. However, 3D imaging of materials is challenging for a number of reasons, including limited field of view, low resolution and difficult sample preparation. Recently, a machine learning method, SliceGAN, was developed to statistically generate 3D microstructural datasets of arbitrary size using a single 2D input slice as training data. In this paper, we present the results from applying SliceGAN to 87 different microstructures, ranging from biological materials to high-strength steels. To demonstrate the accuracy of the synthetic volumes created by SliceGAN, we compare three microstructural properties between the 2D training data and 3D generations, which show good agreement. This new microstructure library both provides valuable 3D microstructures that can be used in models, and also demonstrates the broad applicability of the SliceGAN algorithm.
Measurement(s) | Micrographs • Phase volume fractions • Two point correlations |
Technology Type(s) | Material characterisation techniques (SEM, EBSD, light) • Computation |
Similar content being viewed by others
Background
Understanding the influence of a material’s microstructure on its performance has led to significant advancements in the field of material science1,2,3. Computational methods have played an important role in this success. For example, finite element analysis can capture complex stress fields during mechanical deformation of structural materials4,5, and electro-chemical modelling can help to explain rate limiting factors during battery discharge6,7. These simulations allow high-throughput exploration of a systems performance under a range of conditions8,9. In many fields, this has enabled massive acceleration of the materials optimisation process compared to experiments alone, and with significantly reduced cost. Importantly, 3D datasets are crucial for many applications where 2D datasets cannot be used to determine key material properties. For example, mechanical deformation, crack propagation and tortuosity are three material characteristics that behave fundamentally differently in 3D compared to 2D.
The fidelity of the 3D microstructural datasets commonly required for physical modelling will influence the simulations reliability. Unfortunately, to the authors knowledge, there are no 3D material databases, with most data instead scattered across the literature. This is likely due to the high cost and technical experience required for 3D imaging techniques, which inhibits free sharing of data. Furthermore, where there is data available, it is commonly of limited resolution and field of view due to the intrinsic 3D imaging constraints of techniques such as focussed ion beam scanning electron microscopy and x-ray tomography10. In comparison, diverse, high resolution 2D micrographs are abundantly available online due to the prevalence of 2D imaging techniques such as light microscopy and scanning electron microscopy. DoITPoMS is one excellent micrograph repository with a broad range of alloys, ceramics, bio-materials and more11. UHCSDB is a similar repository, focused solely on high carbon steels12. ASM International has a collection of 4100 micrographs, though access costs a $250 yearly subscription13.
In this paper, we aim to address the disparity between the availability of 2D micrographs compared to 3D. A number of previous approaches have been developed to address this problem through dimensionality expansion, which commonly entails statistical generation of 3D micrographs using statistics from a 2D training image. These are typically physic based and require the extraction of particular metrics from the training data for comparison. For example, sphere packing models using 2D particle size distributions14, poly-crystalline grain growth algorithms15, and data fusion approaches16.
In this work, we use SliceGAN, a recently developed convolutional machine learning algorithm for dimensionality expansion17. A typical GAN uses two convolutional networks (generator and discriminator) to learn to mimic dataset distributions. The generator synthesises fake examples, and the discriminator identifies differences between these fake samples and the true training data distribution. Through iterative learning, the discriminator informs the generator how to make increasingly realistic samples that match the real training data. Importantly, in a typical set-up, the dimensionality of the generated images and the training data match. To facilitate different dimensionalities, SliceGAN uses a simple modification; a 3D generator network produces a sample volume, then a 2D discriminator checks the fidelity of one slice at a time, where the 2D dimensionality of the slice now matches the 2D dimensionality of the training images. The algorithm is described in full in the original manuscript17. SliceGAN is particularly well suited to the task at hand due to a number of key features. First, broad applicability means that the same algorithm and hyper-parameters can be used for a very diverse set of microstructures, as demonstrated in this dataset. Second, high speed training (typically 3 hrs on an RTX6000 GPU) and generation (<3 seconds for a 5003 voxel volume) enables the synthesis of hundreds of large samples for statistical experiments, as well as the generation of volumes far larger than it is currently possible to obtain directly through imaging (>20003 voxel). Third, complete automation of the 2D to 3D algorithm is possible with no user defined inputs, such as statistical features, being required. This combination of strengths makes SliceGAN an excellent candidate for building the first large scale 3D microstructural database from existing open-source 2D data.
The benefits of this database are twofold. First, we provide a diverse 3D microstructural dataset which can be used by the material science community for modelling purposes. Crucially, users are not limited to the single example cube we provide, as each data entry also has an associated trained generator neural network (45 Mb in size) available to download. This can be used to synthesise arbitrary size datasets by cloning the SliceGAN repo and running the relevant scripts (see methods). The second important function of this database is as a demonstration to the material science community of the strengths of SliceGAN. The entries we provide are diverse in their nature, and contained in an easily searchable website. Interested researchers can thus use this website to check whether SliceGAN works on materials in their research field, and see examples of generated outputs. This encourages the submission of more entries to the database, and the further use of SliceGAN in the field of computational materials. The key data processing steps and datasets are presented in Fig. 1.
Methods
As shown in Fig. 1, the database construction required several distinct steps. First, a subset of micrographs were selected using a set of exclusion criteria. A number of simple pre-processing operations were then applied to ensure suitability for the SliceGAN workflow. An automated in-painting method was used to remove scale bars from the micrographs; compared to a crop** approach, this saves crucial data in an already extremely data-scarce setting. Finally, the resulting micrographs are used to train SliceGAN generators, each of which was used to generate an example 3203 cubic volume. Each of these steps can be reproduced by cloning the MicroLib repository and running main.py in the relevant modes, as described in the repository README.
Exclusion criteria
DoITPoMS includes 818 diverse micrographs which can easily be downloaded directly from their website. However, not all are suitable for SliceGAN, which has a number of limitations. As such, the following exclusion criteria are applied to leave 87 feasible microstructures:
-
1.
Microstructure isotropy–SliceGAN can be used for some anisotropic microstructures, but this mode requires multiple perpendicular micrographs which are not available from DoITPoMS.
-
2.
Feature Representativity–SliceGAN relies on feature statistics to generate realistic 3D volumes. Thus, a micrograph containing, for example, a single crystalline grain, is insufficient for the reconstruction of a 3D crystalline microstructure.
-
3.
Even exposure–if parts of the micrograph are brighter than others, this creates significant issues in the final 3D volume, as the algorithm assumes homogeneity.
-
4.
Uniqueness–In some cases there are several replicas of a similar microstructure; Different regions of the same material are always excluded, whilst where there are multiple magnifications, the max and min mag are used to capture different size features.
-
5.
Image quality–Some micrographs are of too low quality to be worth reconstructing.
Image processing
This subsection of images were cropped to remove any borders or non-data regions, such as magnification information underneath the micrograph. Furthermore, of the 87 micrographs, 78 were identified as appropriate for segmentation as they contained easily distinguishable phases. Segmented images are preferable as most material simulation techniques require n-phase datasets such that phase properties can be assigned to a voxel. A simple thresholding process was applied to give n-phase micrographs, which are also better for the SliceGAN training process due to their simplicity. The remaining 9 micrographs were processed as grayscale images. Finally, of the 87 microstructures, 79 had scale bars partially covering the micrograph. In these cases, the colour and location of the scale bar is identified and stored as an annotation to allow in-painting as described in the next section.
Scale bar inpainting
Leaving scale bars in the training data images would result in SliceGAN producing unrealistic features in the generated 3D volume, as it would interpret these objects as microstructural features. The simplest alternative is to entirely crop the region of the micrographs that contain the scale bar; however, this would result in a mean loss of 21% of the training data (when then scale bar and label only actually conceal 1.4% of the image on average). The quality of the final reconstructions could be significantly reduced due to a less representative and diverse distribution of features, which can lead to over-fitting and non-realistic microstructures. To avoid this scenario, we used a machine learning based in-painting technique to remove the scale bars, while leaving all surrounding data untouched18. The locations of the scale bars are identified using a simple gui which allows the user to select the scale bar colour and adjust a threshold until a sufficiently accurate mask is defined. A GAN is then trained to in-paint the masked image, as described in source. The resulting homogeneous microstructure is saved in the intermediate 2D dataset.
SliceGAN 3D reconstruction
Each microstructure is trained on randomly initialised SliceGAN networks for 5000 generator iterations, each with batch size 32, which takes less than 3 hours per entry using an RTX6000 GPU and x CPU. Hyper-parameters are kept consistent with the original SliceGAN paper. The resulting trained generators are saved and then used to synthesise a 320 voxel cube, which takes less than 3 seconds. Figure 2 depicts the outputs at each step for a selection of microstructures.
A selection of representative microstructures are shown, where each row depicts a micrograph, as well as each key stage of data processing required followed by the final 3D volume. Note the red masked scale bars in column 2 have been thickened by 1 pixel in each direction after thresholding of column 1. This ensures that all of the scale bar is in-painted, as edges are sometimes missed due to pixelization.
Data Record
The full dataset can be accessed at zenodo (https://doi.org/10.5281/zenodo.711855919). The dataset is compressed into a single zip file, which contains one sub-folder for each of the 87 entries selected from DoITPoMs. Sub-folder names are derived from the DoITPoMS microstructure ID. Each contains media files (.png,.gif,.mp4) that illustrate the steps taken to generate the data and display examples of resulting microstructures. They also contain trained models and a parameters file (.pt,.data) to allow users to synthesise new volumes.
As well as this set of sub-folders, the root directory also contains an annotations file, data_anns.json. This json contains web-scraped descriptors of each microstructure (with the exception of data_type, which was defined during this study). The full directory tree and brief descriptions of each file are given below.
Besides zenodo, we have also created microlib.io, a website where users can visualise microstructures and search through the microlib database using keywords and filters. Individual media and model files can also be directly downloaded. The web app also provides an easy to use API which the user can query using the same search functionality as the website, allowing for programmatic access to the same data and metadata provided by the frontend. Figure 3 shows a selection of the 3D tifs available, though readers are encouraged to view the MicroLib website for the best visualisation experience.
Technical Validation
Unlike some machine learning methods, such as auto-encoders, GANs do not attempt to exactly recreate images from the training set. Instead, they capture the underlying probability distribution of the training data and synthesise samples with the same distribution of features. This means that there is no ground truth against which the generated outputs can be compared. Thus, to quantify the accuracy of the generated 3D volumes compared to their original 2D training data, we calculate and compare a number of statistical material properties. These tests are only performed on the 78 n-phase materials, as the properties cannot be calculated for grayscale images. First, volume fraction (vf), which is simply the proportion of the voxels assigned to a particular phase. As shown in Fig. 4a), the 3D agrees well with the 2D training data; the mean percentage error, calculated as \(\frac{| \,{\rm{vf(2D)}}-{\rm{vf(3D)}}\,| }{{\rm{vf(2D)}}}\), is 4.7%. Figure 4b) shows a similar comparison for normalised surface area density, which is calculated as the proportion of voxel faces touching both phase 1 and phase 2. The mean percentage error is 4.3%.
Property comparison for 2D training data vs 3D generated volumes. Plots are ordered by increasing value in the 2D dataset (volume fraction and surface area for (a) and (b) respectively. Plots (b) and (c) show the two-point correlation function for four randomly selected microstructures each. Within each plot, only lines of the same colour should be compared as they are from the same material.
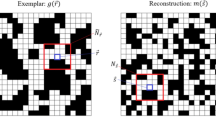
As well as these simple metrics, we also can compare the two-point correlation function (2PC) of the training data and generated volumes. The 2PC gives the probability of finding the same phase pixel at a given pixel separation distance, as describe in more detail in the original SliceGAN paper17. Figure 4c,d each show the 2PC of 8 randomly selected microstructure entries. In general, the curves are very similar for 2D and 3D.
Although the majority of samples show excellent agreement between 2D and 3D,there are a number of outliers, in particular for volume fraction. 5 samples (tags 1, 60, 372, 612, 782) show a volume fraction error greater than 5% (Supplementary Fig. S1 shows three of these microstructures, which exemplify the key failure modes to be discussed below). Notably, sample 60 is also the outlier in the surface area density plot. To explore the nature of this error, three repeats were run on the microstructure to test whether the generator would reproduce the observed behaviour. The new generators gave the same metrics to within 1%. This implies that in this particular use case, well trained generators produce higher volume fractions in 3D than in 2D.
Observing the microstructure itself gives some indication of why this might occur. Sample 60 consists mostly of ovals with a few circles, all of similar sizes. Under the assumption of isotropy, we can ask what 3D structure we expect SliceGAN to generate, and indeed we quickly conclude there is no feasible isotropic 3D volume that can be made where all 2D slices contain only these features. Crucially, we are missing smaller ovals or circles that would be present at the edges of spherical or ellipsoidal features. SliceGAN is thus forced to compromise between accurately reproducing volume fraction versus the exact feature distribution, as both are not possible. Sample 782 suffers from the same problem, whilst sample 1 and 372 have large non-representative features which lead to a similar scenario. Finally, sample 612 is simply poorly segmented. This demonstrates that poor agreement of metrics is one indicator that a non-representative, anisotropic or low quality 2D microstructure has been used to train SliceGAN, which is potentially useful in catching cases where the exclusion criteria were insufficient. However, it is worth noting that unrealistic features might still occur even when volume fractions and two-point correlations match the 2D dataset well. As such, great care should be taken when using these results, and where possible, users of SliceGAN should always compare 2D and 3D statistics for properties relevant to the simulations they are conducting.
Code availability
All code for generating the datasets, including image scra**, preprocessing, inpainting and SliceGAN, can be accessed openly at the MicroLib github repository, https://github.com/tldr-group/microlib, which includes in-depth instruction to ensure reproducibility.
Change history
23 November 2022
In this article the hyperlink provided for microlib.io in the sentence beginning ‘Besides zenodo, we have also created...’ was incorrect, the hyperlink provided for API in the sentence ‘The web app also provides…’ was incorrect, and the hyperlink provided for MicroLib in the sentence ‘Each of these steps…’ was incorrect. The original article has been corrected.
References
Gamble, S. Fabrication-microstructure-performance relationships of reversible solid oxide fuel cell electrodes-review. Materials Science and Technology 27, 1485–1497 (2011).
Plaut, R. L., Herrera, C., Escriba, D. M., Rios, P. R. & Padilha, A. F. A short review on wrought austenitic stainless steels at high temperatures: processing, microstructure, properties and performance. Materials Research 10, 453–460 (2007).
Song, B. et al. Differences in microstructure and properties between selective laser melting and traditional manufacturing for fabrication of metal parts: A review. Frontiers of Mechanical Engineering 10, 111–125 (2015).
Makarem, F. S. & Abed, F. Nonlinear finite element modeling of dynamic localizations in high strength steel columns under impact. International Journal of Impact Engineering 52, 47–61 (2013).
Ma, J., Kong, F. & Kovacevic, R. Finite-element thermal analysis of laser welding of galvanized high-strength steel in a zero-gap lap joint configuration and its experimental verification. Materials & Design (1980–2015) 36, 348–358 (2012).
Wang, Z., Ma, J. & Zhang, L. Finite element thermal model and simulation for a cylindrical li-ion battery. IEEE Access 5, 15372–15379 (2017).
Zadin, V., Kasemägi, H., Aabloo, A. & Brandell, D. Modelling electrode material utilization in the trench model 3d-microbattery by finite element analysis. Journal of Power Sources 195, 6218–6224 (2010).
Liu, S., Zhu, H., Peng, G., Yin, J. & Zeng, X. Microstructure prediction of selective laser melting alsi10mg using finite element analysis. Materials & Design 142, 319–328 (2018).
Prabu, S. B. & Karunamoorthy, L. Microstructure-based finite element analysis of failure prediction in particle-reinforced metal-matrix composite. Journal of materials processing technology 207, 53–62 (2008).
Cocco, A. P. et al. Three-dimensional microstructural imaging methods for energy materials. Physical Chemistry Chemical Physics 15, 16377–16407 (2013).
Elliot, J. DoITPoMS micrograph library. https://www.doitpoms.ac.uk/index.php (2000).
DeCost, B. L., Francis, T. & Holm, E. A. Exploring the microstructure manifold: image texture representations applied to ultrahigh carbon steel microstructures. Acta Materialia 133, 30–40 (2017).
Lupulescu, A., Flowers, T., Vermillion, L. & Henry, S. Asm micrograph databaseâ„¢. Metallography, Microstructure, and Analysis 4 (2015).
Jodrey, W. & Tory, E. Computer simulation of close random packing of equal spheres. Physical review A 32, 2347 (1985).
Groeber, M. A. & Jackson, M. A. Dream. 3d: a digital representation environment for the analysis of microstructure in 3d. Integrating materials and manufacturing innovation 3, 5 (2014).
Xu, H., Usseglio-Viretta, F., Kench, S., Cooper, S. J. & Finegan, D. P. Microstructure reconstruction of battery polymer separators by fusing 2d and 3d image data for transport property analysis. Journal of Power Sources 480, 229101 (2020).
Kench, S. & Cooper, S. J. Generating three-dimensional structures from a two-dimensional slice with generative adversarial network-based dimensionality expansion. Nature Machine Intelligence 3, 299–305 (2021).
Squires, I., Cooper, S. J., Dahari, A. & Kench, S. Two approaches to inpainting microstructure with deep convolutional generative adversarial networks. Arxiv (2022)
Kench, S., Squires, I., Dahari, A. & Cooper, S. J. Microlib dataset. zenodo https://doi.org/10.5281/zenodo.7118559 (2022).
Acknowledgements
This work was supported by funding from the EPSRC Faraday Institution Multi-Scale Modelling project (https://faraday.ac.uk/; EP/S003053/1, grant number FIRG003 received by SK). The authors would also like to thank the authors of DoITPoMS for their open source dataset, without which this work would not be possible.
Author information
Authors and Affiliations
Contributions
S.K. developed the code required for the generation of the 2D dataset, performed the technical validation and wrote the paper. A.D. ran SliceGAN scripts on the Imperial College high-performance to generate 3D volumes. I.S. developed the microlib.io website. All authors contributed to the conceptualisation of the project and editing of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kench, S., Squires, I., Dahari, A. et al. MicroLib: A library of 3D microstructures generated from 2D micrographs using SliceGAN. Sci Data 9, 645 (2022). https://doi.org/10.1038/s41597-022-01744-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-022-01744-1
- Springer Nature Limited
This article is cited by
-
Generative adversarial network (GAN) enabled Statistically equivalent virtual microstructures (SEVM) for modeling cold spray formed bimodal polycrystals
npj Computational Materials (2024)
-
Generating 3D images of material microstructures from a single 2D image: a denoising diffusion approach
Scientific Reports (2024)
-
MICRO2D: A Large, Statistically Diverse, Heterogeneous Microstructure Dataset
Integrating Materials and Manufacturing Innovation (2024)