Abstract
For the past three decades, apolipoprotein E (APOE) has been known as the single greatest genetic modulator of sporadic Alzheimer disease (AD) risk, influencing both the average age of onset and the lifetime risk of develo** AD. The APOEε4 allele significantly increases AD risk, whereas the ε2 allele is protective relative to the most common ε3 allele. However, large differences in effect size exist across ethnoracial groups that are likely to depend on both global genetic ancestry and local genetic ancestry, as well as gene–environment interactions. Although early studies linked APOE to amyloid-β — one of the two culprit aggregation-prone proteins that define AD — in the past decade, mounting work has associated APOE with other neurodegenerative proteinopathies and broader ageing-related brain changes, such as neuroinflammation, energy metabolism failure, loss of myelin integrity and increased blood–brain barrier permeability, with potential implications for longevity and resilience to pathological protein aggregates. Novel mouse models and other technological advances have also enabled a number of therapeutic approaches aimed at either attenuating the APOEε4-linked increased AD risk or enhancing the APOEε2-linked AD protection. This Review summarizes this progress and highlights areas for future research towards the development of APOE-directed therapeutics.
Key points
-
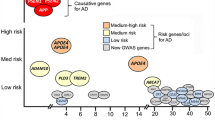
The risk of Alzheimer disease associated with the APOE genotype is modulated by global and local genetic ancestries, other genetic risk loci and the lifetime exposome of an individual.
-
APOE missense mutations are providing key insights into the pathophysiology of the classic three APOE isoforms.
-
The APOE genotype might modulate the risk of other neurodegenerative diseases by influencing the pathobiology of their culprit aggregation-prone proteins.
-
The APOE isoforms affect a wide range of molecular and cellular functions in multiple brain cell types via cell-autonomous and non-autonomous mechanisms.
-
Several strategies to target APOE therapeutically have shown efficacy in preclinical studies and hold promise for translation into clinical trials.



Similar content being viewed by others
References
Glenner, G. G. & Wong, C. W. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885–890 (1984).
van Dyck, C. H. et al. Lecanemab in early Alzheimer’s disease. N. Engl. J. Med. 388, 9–21 (2023).
Mintun, M. A. et al. Donanemab in early Alzheimer’s disease. N. Engl. J. Med. 384, 1691–1704 (2021).
Serrano-Pozo, A., Aldridge, G. M. & Zhang, Q. Four decades of research in Alzheimer’s disease (1975–2014): a bibliometric and scientometric analysis. J. Alzheimers Dis. 59, 763–783 (2017).
Corder, E. H. et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923 (1993).
Farrer, L. A. et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278, 1349–1356 (1997).
Corder, E. H. et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet. 7, 180–184 (1994).
Reiman, E. M. et al. Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat. Commun. 11, 667 (2020).
Mattsson, N. et al. Prevalence of the apolipoprotein E ε4 allele in amyloid β positive subjects across the spectrum of Alzheimer’s disease. Alzheimers Dement. 14, 913–924 (2018).
Belloy, M. E. et al. APOE genotype and Alzheimer disease risk across age, sex, and population ancestry. JAMA Neurol. 80, 1284–1294 (2023).
Granot-Hershkovitz, E. et al. APOE alleles’ association with cognitive function differs across Hispanic/Latino groups and genetic ancestry in the study of Latinos — investigation of neurocognitive aging (HCHS/SOL). Alzheimers Dement. 17, 466–474 (2021).
Rajabli, F. et al. Ancestral origin of ApoE ε4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS Genet. 14, e1007791 (2018).
Blue, E. E., Horimoto, A. R. V. R., Mukherjee, S., Wijsman, E. M. & Thornton, T. A. Local ancestry at APOE modifies Alzheimer’s disease risk in Caribbean Hispanics. Alzheimers Dement. 15, 1524–1532 (2019).
Griswold, A. J. et al. Increased APOE ε4 expression is associated with the difference in Alzheimer’s disease risk from diverse ancestral backgrounds. Alzheimers Dement. 17, 1179–1188 (2021).
Nuytemans, K. et al. Identifying differential regulatory control of APOE ɛ4 on African versus European haplotypes as potential therapeutic targets. Alzheimers Dement. 18, 1930–1942 (2022).
Fortea, J. et al. APOE4 homozygosity represents a distinct genetic form of Alzheimer’s disease. Nat. Med. 30, 1284–1291 (2024).
Qian, J. et al. APOE-related risk of mild cognitive impairment and dementia for prevention trials: an analysis of four cohorts. PLoS Med. 14, e1002254 (2017).
Stites, S. D. et al. Patients asking about APOE gene test results? Here’s what to tell them. J. Fam. Pract. 71, E1–E7 (2022).
Wightman, D. P. et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 53, 1276–1282 (2021).
Bellenguez, C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022).
Huq, A. J. et al. Polygenic score modifies risk for Alzheimer’s disease in APOE ε4 homozygotes at phenotypic extremes. Alzheimers Dement. 13, e12226 (2021).
Ebenau, J. L. et al. Risk of dementia in APOE ε4 carriers is mitigated by a polygenic risk score. Alzheimers Dement. 13, e12229 (2021).
Erickson, C. M. et al. KLOTHO heterozygosity attenuates APOE4-related amyloid burden in preclinical AD. Neurology 92, e1878–e1889 (2019).
Belloy, M. E. et al. Association of klotho vs heterozygosity with risk of Alzheimer disease in individuals who carry APOE4. JAMA Neurol. 77, 849–862 (2020).
Neitzel, J. et al. KL-VS heterozygosity is associated with lower amyloid-dependent tau accumulation and memory impairment in Alzheimer’s disease. Nat. Commun. 12, 3825 (2021).
Ali, M. et al. Leveraging large multi-center cohorts of Alzheimer disease endophenotypes to understand the role of klotho heterozygosity on disease risk. PLoS ONE 17, e0267298 (2022).
Huq, A. J. et al. Genetic resilience to Alzheimer’s disease in APOE ε4 homozygotes: a systematic review. Alzheimers Dement. 15, 1612–1623 (2019).
Serrano-Pozo, A. & Growdon, J. H. Is Alzheimer’s disease risk modifiable? J. Alzheimers Dis. 67, 795–819 (2019).
Jaisa-Aad, M., Muñoz-Castro, C. & Serrano-Pozo, A. Update on modifiable risk factors for Alzheimer’s disease and related dementias. Curr. Opin. Neurol. 37, 166–181 (2024).
Kolli, A. et al. Interactions between the apolipoprotein E4 gene and modifiable risk factors for cognitive impairment: a nationally representative panel study. BMC Geriatr. 22, 938 (2022).
Langella, S. et al. Effect of apolipoprotein genotype and educational attainment on cognitive function in autosomal dominant Alzheimer’s disease. Nat. Commun. 14, 5120 (2023).
Park, S.-Y. et al. Modifying effects of race and ethnicity and APOE on the association of physical activity with risk of Alzheimer’s disease and related dementias. Alzheimers Dement. 19, 507–517 (2023).
Jia, J. et al. Association between healthy lifestyle and memory decline in older adults: 10 year, population based, prospective cohort study. Br. Med. J. 380, e072691 (2023).
Park, S.-Y. et al. Racial and ethnic differences in the population-attributable fractions of Alzheimer disease and related dementias. Neurology 102, e208116 (2024).
Lee, M. et al. Variation in population attributable fraction of dementia associated with potentially modifiable risk factors by race and ethnicity in the US. JAMA Netw. Open 5, e2219672 (2022).
Arboleda-Velasquez, J. F. et al. Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: a case report. Nat. Med. 25, 1680–1683 (2019).
He, K. Y. et al. Characterization of APOE Christchurch carriers in 455,306 UK Biobank participants. Mol. Neurodegener. 18, 92 (2023).
Wardell, M. R., Brennan, S. O., Janus, E. D., Fraser, R. & Carrell, R. W. Apolipoprotein E2-Christchurch (136 Arg–Ser). New variant of human apolipoprotein E in a patient with type III hyperlipoproteinemia. J. Clin. Invest. 80, 483–490 (1987).
Pocovi, M. et al. Incomplete dominance of type III hyperlipoproteinemia is associated with the rare apolipoprotein E2 (Arg136→Ser) variant in multigenerational pedigree studies. Atherosclerosis 122, 33–46 (1996).
Hernandez, I. et al. Heterozygous APOE Christchurch in familial Alzheimer’s disease without mutations in other Mendelian genes. Neuropathol. Appl. Neurobiol. 47, 579–582 (2021).
Le Guen, Y. et al. Association of African ancestry-specific APOE missense variant r145c with risk of Alzheimer disease. JAMA 329, 551–560 (2023).
Medway, C. W. et al. ApoE variant p.V236E is associated with markedly reduced risk of Alzheimer’s disease. Mol. Neurodegener. 9, 11 (2014).
Le Guen, Y. et al. Association of rare APOE missense variants V236E and R251G with risk of Alzheimer disease. JAMA Neurol. 79, 652–663 (2022).
Liu, C.-C. et al. APOE3-Jacksonville (V236E) variant reduces self-aggregation and risk of dementia. Sci. Transl. Med. 13, eabc9375 (2021).
Boyle, P. A. et al. Person-specific contribution of neuropathologies to cognitive loss in old age. Ann. Neurol. 83, 74–83 (2018).
Bennett, D. A. et al. Amyloid mediates the association of apolipoprotein E e4 allele to cognitive function in older people. J. Neurol. Neurosurg. Psychiatry 76, 1194–1199 (2005).
Mungas, D., Tractenberg, R., Schneider, J. A., Crane, P. K. & Bennett, D. A. A 2-process model for neuropathology of Alzheimer’s disease. Neurobiol. Aging 35, 301–308 (2014).
Serrano-Pozo, A., Qian, J., Monsell, S. E., Betensky, R. A. & Hyman, B. T. APOEε2 is associated with milder clinical and pathological Alzheimer disease. Ann. Neurol. 77, 917–929 (2015).
Goldberg, T. E., Huey, E. D. & Devanand, D. P. Association of APOE ε2 genotype with Alzheimer’s and non-Alzheimer’s neurodegenerative pathologies. Nat. Commun. 11, 4727 (2020).
Goldberg, T. E., Huey, E. D. & Devanand, D. P. Associations of APOE ε2 genotype with cerebrovascular pathology: a postmortem study of 1275 brains. J. Neurol. Neurosurg. Psychiatry https://doi.org/10.1136/jnnp-2020-323746 (2020).
Yu, L. et al. APOE and cerebral amyloid angiopathy in community-dwelling older persons. Neurobiol. Aging 36, 2946–2953 (2015).
Thal, D. R. et al. Capillary cerebral amyloid angiopathy identifies a distinct APOE ε4-associated subtype of sporadic Alzheimer’s disease. Acta Neuropathol. 120, 169–183 (2010).
Farfel, J. M., Yu, L., De Jager, P. L., Schneider, J. A. & Bennett, D. A. Association of APOE with tau-tangle pathology with and without β-amyloid. Neurobiol. Aging 37, 19–25 (2016).
Oveisgharan, S. et al. APOE ε2ε4 genotype, incident AD and MCI, cognitive decline, and AD pathology in older adults. Neurology 90, e2127–e2134 (2018).
Lamar, M. et al. APOE genotypes as a risk factor for age-dependent accumulation of cerebrovascular disease in older adults. Alzheimers Dement. 15, 258–266 (2019).
Greenberg, S. M. et al. Association of apolipoprotein E ε2 and vasculopathy in cerebral amyloid angiopathy. Neurology 50, 961–965 (1998).
McCarron, M. O. et al. The apolipoprotein E ε2 allele and the pathological features in cerebral amyloid angiopathy-related hemorrhage. J. Neuropathol. Exp. Neurol. 58, 711–718 (1999).
Serrano-Pozo, A. et al. Examination of the clinicopathologic continuum of Alzheimer disease in the autopsy cohort of the National Alzheimer Coordinating Center. J. Neuropathol. Exp. Neurol. 72, 1182–1192 (2013).
Robinson, J. L. et al. The development and convergence of co-pathologies in Alzheimer’s disease. Brain 144, 953–962 (2021).
Karanth, S. et al. Prevalence and clinical phenotype of quadruple misfolded proteins in older adults. JAMA Neurol. 77, 1299–1307 (2020).
Walker, J. M. & Richardson, T. E. Cognitive resistance to and resilience against multiple comorbid neurodegenerative pathologies and the impact of APOE status. J. Neuropathol. Exp. Neurol. 82, 110–119 (2023).
Dickson, D. W. et al. APOE ε4 is associated with severity of Lewy body pathology independent of Alzheimer pathology. Neurology 91, e1182–e1195 (2018).
Yang, H.-S. et al. Evaluation of TDP-43 proteinopathy and hippocampal sclerosis in relation to APOE ε4 haplotype status: a community-based cohort study. Lancet Neurol. 17, 773–781 (2018).
Wennberg, A. M. et al. Association of apolipoprotein E ε4 with transactive response DNA-binding protein 43. JAMA Neurol. 75, 1347–1354 (2018).
Robinson, J. L. et al. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain 141, 2181–2193 (2018).
Zhao, N. et al. APOE ε2 is associated with increased tau pathology in primary tauopathy. Nat. Commun. 9, 4388 (2018).
Sabir, M. S. et al. Assessment of APOE in atypical parkinsonism syndromes. Neurobiol. Dis. 127, 142–146 (2019).
Atherton, K. et al. Association of APOE genotypes and chronic traumatic encephalopathy. JAMA Neurol. 79, 787–796 (2022).
Davis, A. A. et al. APOE genotype regulates pathology and disease progression in synucleinopathy. Sci. Transl. Med. 12, eaay3069 (2020).
Tsuang, D. et al. APOE ε4 increases risk for dementia in pure synucleinopathies. JAMA Neurol. 70, 223–228 (2013).
Zhao, N. et al. APOE4 exacerbates α-synuclein pathology and related toxicity independent of amyloid. Sci. Transl. Med. 12, eaay1809 (2020).
Kaivola, K., Shah, Z. & Chia, R., International LBD Genomics Consortium & Scholz, S. W. Genetic evaluation of dementia with Lewy bodies implicates distinct disease subgroups. Brain 145, 1757–1762 (2022).
Talyansky, S., Le Guen, Y., Kasireddy, N., Belloy, M. E. & Greicius, M. D. APOE-ε4 and BIN1 increase risk of Alzheimer’s disease pathology but not specifically of Lewy body pathology. Acta Neuropathol. Commun. 11, 149 (2023).
Ogaki, K. et al. Multiple system atrophy and apolipoprotein E. Mov. Disord. 33, 647–650 (2018).
Meneses, A. D. et al. APOE2 exacerbates TDP-43 related toxicity in the absence of Alzheimer pathology. Ann. Neurol. 93, 830–843 (2023).
Caselli, R. J. et al. Longitudinal modeling of age-related memory decline and the APOE ε4 effect. N. Engl. J. Med. 361, 255–263 (2009).
Shinohara, M. et al. APOE2 eases cognitive decline during aging: clinical and preclinical evaluations. Ann. Neurol. 79, 758–774 (2016).
Yu, L. et al. APOE ε4, Alzheimer’s disease pathology, cerebrovascular disease, and cognitive change over the years prior to death. Psychol. Aging 28, 1015–1023 (2013).
Nichols, E. et al. AD and non-AD mediators of the pathway between the APOE genotype and cognition. Alzheimers Dement. 19, 2508–2519 (2023).
Qian, J., Betensky, R. A., Hyman, B. T. & Serrano-Pozo, A. Association of APOE genotype with heterogeneity of cognitive decline rate in Alzheimer disease. Neurology 96, e2414–e2428 (2021).
Qian, J., Zhang, Y., Betensky, R. A., Hyman, B. T. & Serrano-Pozo, A. Neuropathology-independent association between APOE genotype and cognitive decline rate in the normal aging-early Alzheimer continuum. Neurol. Genet. 9, e200055 (2023).
Bejanin, A. et al. Association of apolipoprotein E ɛ4 allele with clinical and multimodal biomarker changes of Alzheimer disease in adults with down syndrome. JAMA Neurol. 78, 937–947 (2021).
Vélez, J. I. et al. APOE*E2 allele delays age of onset in PSEN1 E280A Alzheimer’s disease. Mol. Psychiatry 21, 916–924 (2016).
Willer, C. J. et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40, 161–169 (2008).
Kathiresan, S. et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40, 189–197 (2008).
Natarajan, P. et al. Multiethnic exome-wide association study of subclinical atherosclerosis. Circ. Cardiovasc. Genet. 9, 511–520 (2016).
Kuo, C.-L., Pilling, L. C., Atkins, J. L., Kuchel, G. A. & Melzer, D. ApoE ε2 and aging-related outcomes in 379,000 UK Biobank participants. Aging 12, 12222–12233 (2020).
Lumsden, A. L., Mulugeta, A., Zhou, A. & Hyppönen, E. Apolipoprotein E (APOE) genotype-associated disease risks: a phenome-wide, registry-based, case–control study utilising the UK Biobank. eBioMedicine 59, 102954 (2020).
Ghiselli, G., Gregg, R. E., Zech, L. A., Schaefer, E. J. & Brewer, H. B. Phenotype study of apolipoprotein E isoforms in hyperlipoproteinaemic patients. Lancet 2, 405–407 (1982).
Joshi, P. K. et al. Genome-wide meta-analysis associates HLA-DQA1/DRB1 and LPA and lifestyle factors with human longevity. Nat. Commun. 8, 910 (2017).
Pilling, L. C. et al. Human longevity: 25 genetic loci associated in 389,166 UK biobank participants. Aging 9, 2504–2520 (2017).
Deelen, J. et al. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat. Commun. 10, 3669 (2019).
Wolters, F. J. et al. The impact of APOE genotype on survival: results of 38,537 participants from six population-based cohorts (E2-CHARGE). PLoS ONE 14, e0219668 (2019).
Shinohara, M. et al. APOE2 is associated with longevity independent of Alzheimer’s disease. eLife 9, e62199 (2020).
Yang, L. G., March, Z. M., Stephenson, R. A. & Narayan, P. S. Apolipoprotein E in lipid metabolism and neurodegenerative disease. Trends Endocrinol. Metab. 34, 430–445 (2023).
Xu, Q. et al. Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J. Neurosci. 26, 4985–4994 (2006).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Hirsch-Reinshagen, V. et al. Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain. J. Biol. Chem. 279, 41197–41207 (2004).
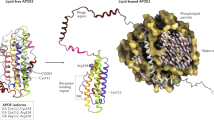
Chen, Y., Strickland, M. R., Soranno, A. & Holtzman, D. M. Apolipoprotein E: structural insights and links to Alzheimer disease pathogenesis. Neuron 109, 205–221 (2021).
Frieden, C., Wang, H. & Ho, C. M. W. A mechanism for lipid binding to apoE and the role of intrinsically disordered regions coupled to domain–domain interactions. Proc. Natl Acad. Sci. USA 114, 6292–6297 (2017).
Strickland, M. R. et al. Apolipoprotein E secreted by astrocytes forms antiparallel dimers in discoidal lipoproteins. Neuron 112, 1100–1109.e5 (2024).
Nguyen, D. et al. Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4. Biochemistry 49, 10881–10889 (2010).
Fernández-Calle, R. et al. APOE in the bullseye of neurodegenerative diseases: impact of the APOE genotype in Alzheimer’s disease pathology and brain diseases. Mol. Neurodegener. 17, 62 (2022).
Kanekiyo, T., Liu, C.-C., Shinohara, M., Li, J. & Bu, G. LRP1 in brain vascular smooth muscle cells mediates local clearance of Alzheimer’s amyloid-β. J. Neurosci. 32, 16458–16465 (2012).
Kanekiyo, T. et al. Neuronal clearance of amyloid-β by endocytic receptor LRP1. J. Neurosci. 33, 19276–19283 (2013).
Shi, Y. et al. Overexpressing low-density lipoprotein receptor reduces tau-associated neurodegeneration in relation to apoE-linked mechanisms. Neuron 109, 2413–2426.e7 (2021).
Castellano, J. M. et al. Low-density lipoprotein receptor overexpression enhances the rate of brain-to-blood Aβ clearance in a mouse model of β-amyloidosis. Proc. Natl Acad. Sci. USA 109, 15502–15507 (2012).
Kim, J. et al. Overexpression of low-density lipoprotein receptor in the brain markedly inhibits amyloid deposition and increases extracellular A beta clearance. Neuron 64, 632–644 (2009).
Tzioras, M., Davies, C., Newman, A., Jackson, R. & Spires-Jones, T. Invited review: APOE at the interface of inflammation, neurodegeneration and pathological protein spread in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 45, 327–346 (2019).
Wisniewski, T. & Frangione, B. Apolipoprotein E: a pathological chaperone protein in patients with cerebral and systemic amyloid. Neurosci. Lett. 135, 235–238 (1992).
Jones, P. B. et al. Apolipoprotein E: isoform specific differences in tertiary structure and interaction with amyloid-β in human Alzheimer brain. PLoS One 6, e14586 (2011).
Liu, C.-C. et al. ApoE4 accelerates early seeding of amyloid pathology. Neuron 96, 1024–1032.e3 (2017).
Hashimoto, T. et al. Apolipoprotein E, especially apolipoprotein E4, increases the oligomerization of amyloid β peptide. J. Neurosci. 32, 15181–15192 (2012).
Hori, Y., Hashimoto, T., Nomoto, H., Hyman, B. T. & Iwatsubo, T. Role of apolipoprotein E in β-amyloidogenesis: isoform-specific effects on protofibril to fibril conversion of Aβ in vitro and brain Aβ deposition in vivo. J. Biol. Chem. 293, 7267 (2018).
Garai, K., Verghese, P. B., Baban, B., Holtzman, D. M. & Frieden, C. The binding of apolipoprotein E to oligomers and fibrils of amyloid-β alters the kinetics of amyloid aggregation. Biochemistry 53, 6323–6331 (2014).
Kara, E. et al. A flow cytometry-based in vitro assay reveals that formation of apolipoprotein E (ApoE)-amyloid beta complexes depends on ApoE isoform and cell type. J. Biol. Chem. 293, 13247–13256 (2018).
Fagan, A. M. et al. Human and murine ApoE markedly alters A beta metabolism before and after plaque formation in a mouse model of Alzheimer’s disease. Neurobiol. Dis. 9, 305–318 (2002).
Bales, K. R. et al. Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer’s disease. Proc. Natl Acad. Sci. USA 96, 15233–15238 (1999).
Irizarry, M. C. et al. Apolipoprotein E affects the amount, form, and anatomical distribution of amyloid beta-peptide deposition in homozygous APP(V717F) transgenic mice. Acta Neuropathol. 100, 451–458 (2000).
Youmans, K. L. et al. APOE4-specific changes in Aβ accumulation in a new transgenic mouse model of Alzheimer disease. J. Biol. Chem. 287, 41774–41786 (2012).
Huang, Y.-W. A., Zhou, B., Wernig, M. & Südhof, T. C. ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription and Aβ secretion. Cell 168, 427–441.e21 (2017).
Hudry, E. et al. Opposing roles of apolipoprotein E in aging and neurodegeneration. Life Sci. Alliance 2, e201900325 (2019).
Mak, A. C. Y. et al. Effects of the absence of apolipoprotein E on lipoproteins, neurocognitive function, and retinal function. JAMA Neurol. 71, 1228 (2014).
Huynh, T.-P. V. et al. Age-dependent effects of apoE reduction using antisense oligonucleotides in a model of β-amyloidosis. Neuron 96, 1013–1023.e4 (2017).
Hou, T. et al. Apolipoprotein E facilitates amyloid-β oligomer-induced tau phosphorylation. J. Alzheimers Dis. 74, 521–534 (2020).
Harris, F. M., Brecht, W. J., Xu, Q., Mahley, R. W. & Huang, Y. Increased tau phosphorylation in apolipoprotein E4 transgenic mice is associated with activation of extracellular signal-regulated kinase: modulation by zinc. J. Biol. Chem. 279, 44795–44801 (2004).
Shi, Y. et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549, 523–527 (2017).
Hudry, E. et al. Gene transfer of human Apoe isoforms results in differential modulation of amyloid deposition and neurotoxicity in mouse brain. Sci. Transl. Med. 5, 212ra161 (2013).
Parhizkar, S. & Holtzman, D. M. APOE mediated neuroinflammation and neurodegeneration in Alzheimer’s disease. Semin. Immunol. 59, 101594 (2022).
Shi, Y. et al. Microglia drive APOE-dependent neurodegeneration in a tauopathy mouse model. J. Exp. Med. 216, 2546–2561 (2019).
**, Y. et al. APOE4 exacerbates α-synuclein seeding activity and contributes to neurotoxicity in Alzheimer’s disease with Lewy body pathology. Acta Neuropathol. 143, 641–662 (2022).
Emamzadeh, F. N., Aojula, H., McHugh, P. C. & Allsop, D. Effects of different isoforms of apoE on aggregation of the α-synuclein protein implicated in Parkinson’s disease. Neurosci. Lett. 618, 146–151 (2016).
Lloyd, G. M. et al. Carboxyl truncation of α-synuclein occurs early and is influenced by human APOE genotype in transgenic mouse models of α-synuclein pathogenesis. Acta Neuropathol. Commun. 11, 119 (2023).
Nemergut, M. et al. Domino-like effect of C112R mutation on ApoE4 aggregation and its reduction by Alzheimer’s disease drug candidate. Mol. Neurodegener. 18, 38 (2023).
Zhang, Y. et al. A monomeric, biologically active, full-length human apolipoprotein E. Biochemistry 46, 10722–10732 (2007).
Garai, K., Baban, B. & Frieden, C. Dissociation of apoE oligomers to monomers is required for high affinity binding to phospholipid vesicles. Biochemistry 50, 2550–2558 (2011).
**ong, M. et al. APOE immunotherapy reduces cerebral amyloid angiopathy and amyloid plaques while improving cerebrovascular function. Sci. Transl. Med. 13, eabd7522 (2021).
Gratuze, M. et al. APOE antibody inhibits Aβ‐associated tau seeding and spreading in a mouse model. Ann. Neurol. 91, 847–852 (2022).
Liao, F. et al. Targeting of nonlipidated, aggregated apoE with antibodies inhibits amyloid accumulation. J. Clin. Invest. 128, 2144–2155 (2018).
Kara, E. et al. Isoform- and cell type-specific structure of apolipoprotein E lipoparticles as revealed by a novel Forster resonance energy transfer assay. J. Biol. Chem. 292, 14720–14729 (2017).
Stuchell-Brereton, M. D. et al. Apolipoprotein E4 has extensive conformational heterogeneity in lipid-free and lipid-bound forms. Proc. Natl Acad. Sci. USA 120, e2215371120 (2023).
Suidan, G. L. & Ramaswamy, G. Targeting apolipoprotein E for Alzheimer’s disease: an industry perspective. Int. J. Mol. Sci. 20, 2161 (2019).
Lin, Y.-T. et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron 98, 1141–1154.e7 (2018).
Tcw, J. et al. Cholesterol and matrisome pathways dysregulated in astrocytes and microglia. Cell 185, 2213–2233.e25 (2022).
Steele, O. G. et al. A multi‐hit hypothesis for an APOE4‐dependent pathophysiological state. Eur. J. Neurosci. 56, 5476–5515 (2022).
Farmer, B. C., Kluemper, J. & Johnson, L. A. Apolipoprotein E4 alters astrocyte fatty acid metabolism and lipid droplet formation. Cells 8, 182 (2019).
Schmukler, E. et al. Altered mitochondrial dynamics and function in APOE4-expressing astrocytes. Cell Death Dis. 11, 578 (2020).
Mahan, T. E. et al. Selective reduction of astrocyte apoE3 and apoE4 strongly reduces Aβ accumulation and plaque-related pathology in a mouse model of amyloidosis. Mol. Neurodegener. 17, 13 (2022).
Wang, C. et al. Selective removal of astrocytic APOE4 strongly protects against tau-mediated neurodegeneration and decreases synaptic phagocytosis by microglia. Neuron https://doi.org/10.1016/j.neuron.2021.03.024 (2021).
Krasemann, S. et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9 (2017).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017).
Serrano-Pozo, A. et al. Effect of APOE alleles on the glial transcriptome in normal aging and Alzheimer’s disease. Nat. Aging 1, 919–931 (2021).
Das, S. et al. Distinct transcriptomic responses to Aβ plaques, neurofibrillary tangles, and APOE in Alzheimer’s disease. Alzheimers Dement. 20, 74–90 (2023).
Stephen, T. L. et al. APOE genotype and sex affect microglial interactions with plaques in Alzheimer’s disease mice. Acta Neuropathol. Commun. 7, 82 (2019).
Wang, Y. et al. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213, 667–675 (2016).
Lanfranco, M. F., Sepulveda, J., Kopetsky, G. & Rebeck, G. W. Expression and secretion of apoE isoforms in astrocytes and microglia during inflammation. Glia 69, 1478–1493 (2021).
Henningfield, C. M., Arreola, M. A., Soni, N., Spangenberg, E. E. & Green, K. N. Microglia-specific ApoE knock-out does not alter Alzheimer’s disease plaque pathogenesis or gene expression. Glia 70, 287–302 (2022).
Yin, Z. et al. APOE4 impairs the microglial response in Alzheimer’s disease by inducing TGFβ-mediated checkpoints. Nat. Immunol. 24, 1839–1853 (2023).
Buttini, M. et al. Cellular source of apolipoprotein E4 determines neuronal susceptibility to excitotoxic injury in transgenic mice. Am. J. Pathol. 177, 563–569 (2010).
Konings, S. C., Torres-Garcia, L., Martinsson, I. & Gouras, G. K. Astrocytic and neuronal apolipoprotein E isoforms differentially affect neuronal excitability. Front. Neurosci. 15, 734001 (2021).
Koutsodendris, N. et al. Neuronal APOE4 removal protects against tau-mediated gliosis, neurodegeneration and myelin deficits. Nat. Aging 3, 275–296 (2023).
Cheng, G. W.-Y. et al. Apolipoprotein E ε4 mediates myelin breakdown by targeting oligodendrocytes in sporadic Alzheimer disease. J. Neuropathol. Exp. Neurol. 81, 717–730 (2022).
Blanchard, J. W. et al. APOE4 impairs myelination via cholesterol dysregulation in oligodendrocytes. Nature 611, 769–779 (2022).
Mok, K. K.-S. et al. Apolipoprotein E ε4 disrupts oligodendrocyte differentiation by interfering with astrocyte-derived lipid transport. J. Neurochem. 165, 55–75 (2023).
Chang, S. et al. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc. Natl Acad. Sci. USA 102, 18694–18699 (2005).
Chen, H.-K. et al. Apolipoprotein E4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. J. Biol. Chem. 286, 5215–5221 (2011).
Parcon, P. A. et al. Apolipoprotein E4 inhibits autophagy gene products through direct, specific binding to CLEAR motifs. Alzheimers Dement. 14, 230–242 (2018).
Mary, A., Eysert, F., Checler, F. & Chami, M. Mitophagy in Alzheimer’s disease: molecular defects and therapeutic approaches. Mol. Psychiatry 28, 202–216 (2023).
Lee, H. et al. ApoE4-dependent lysosomal cholesterol accumulation impairs mitochondrial homeostasis and oxidative phosphorylation in human astrocytes. Cell Rep. 42, 113183 (2023).
Yin, J. et al. Effect of ApoE isoforms on mitochondria in Alzheimer disease. Neurology 94, e2404–e2411 (2020).
Calvo-Rodriguez, M. et al. Increased mitochondrial calcium levels associated with neuronal death in a mouse model of Alzheimer’s disease. Nat. Commun. 11, 2146 (2020).
Calvo-Rodriguez, M. et al. Real-time imaging of mitochondrial redox reveals increased mitochondrial oxidative stress associated with amyloid β aggregates in vivo in a mouse model of Alzheimer’s disease. Mol. Neurodegener. 19, 6 (2024).
Orr, A. L. et al. Neuronal apolipoprotein E4 expression results in proteome-wide alterations and compromises bioenergetic capacity by disrupting mitochondrial function. J. Alzheimers Dis. 68, 991–1011 (2019).
Area-Gomez, E. et al. APOE4 is associated with differential regional vulnerability to bioenergetic deficits in aged APOE mice. Sci. Rep. 10, 4277 (2020).
Williams, H. C. et al. APOE alters glucose flux through central carbon pathways in astrocytes. Neurobiol. Dis. 136, 104742 (2020).
Lee, S. et al. APOE modulates microglial immunometabolism in response to age, amyloid pathology, and inflammatory challenge. Cell Rep. 42, 112196 (2023).
Qi, G. et al. ApoE4 impairs neuron–astrocyte coupling of fatty acid metabolism. Cell Rep. 34, 108572 (2021).
Haney, M. S. et al. APOE4/4 is linked to damaging lipid droplets in Alzheimer’s disease microglia. Nature 628, 154–161 (2024).
Foley, P. Lipids in Alzheimer’s disease: a century-old story. Biochim. Biophys. Acta 1801, 750–753 (2010).
Wynne, M. E. et al. APOE expression and secretion are modulated by mitochondrial dysfunction. eLife 12, e85779 (2023).
Huynh, T.-P. V. et al. Lack of hepatic apoE does not influence early Aβ deposition: observations from a new APOE knock-in model. Mol. Neurodegener. 14, 37 (2019).
Linton, M. F. et al. Phenotypes of apolipoprotein B and apolipoprotein E after liver transplantation. J. Clin. Invest. 88, 270–281 (1991).
Bell, R. D. et al. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature 485, 512–516 (2012).
Jackson, R. J. et al. APOE4 derived from astrocytes leads to blood–brain barrier impairment. Brain 145, 3582–3593 (2022).
Montagne, A. et al. APOE4 leads to blood–brain barrier dysfunction predicting cognitive decline. Nature 581, 71–76 (2020).
**ong, M. et al. Astrocytic APOE4 removal confers cerebrovascular protection despite increased cerebral amyloid angiopathy. Mol. Neurodegener. 18, 17 (2023).
Bonnar, O. et al. APOE4 expression confers a mild, persistent reduction in neurovascular function in the visual cortex and hippocampus of awake mice. J. Cereb. Blood Flow Metab. 43, 1826–1841 (2023).
Koizumi, K. et al. Apoε4 disrupts neurovascular regulation and undermines white matter integrity and cognitive function. Nat. Commun. 9, 3816 (2018).
Wiesmann, M. et al. A dietary treatment improves cerebral blood flow and brain connectivity in aging apoE4 mice. Neural Plast. 2016, 6846721 (2016).
Cummings, J. et al. Lecanemab: appropriate use recommendations. J. Prev. Alzheimers Dis. 10, 362–377 (2023).
Vance, J. M. et al. Report of the APOE4 National Institute on Aging/Alzheimer Disease Sequencing Project Consortium Working Group: reducing APOE4 in carriers is a therapeutic goal for Alzheimer’s disease. Ann. Neurol. 95, 625–634 (2024).
Koffie, R. M. et al. Apolipoprotein E4 effects in Alzheimer’s disease are mediated by synaptotoxic oligomeric amyloid-β. Brain 135, 2155–2168 (2012).
Kuszczyk, M. A. et al. Blocking the interaction between apolipoprotein E and Aβ reduces intraneuronal accumulation of Aβ and inhibits synaptic degeneration. Am. J. Pathol. 182, 1750–1768 (2013).
Liu, S. et al. Blocking the apolipoprotein E/amyloid β interaction in triple transgenic mice ameliorates Alzheimer’s disease related amyloid β and tau pathology. J. Neurochem. 128, 577–591 (2014).
Christensen, D. J. et al. Apolipoprotein E and peptide mimetics modulate inflammation by binding the SET protein and activating protein phosphatase 2A. J. Immunol. 186, 2535–2542 (2011).
Krishnamurthy, K. et al. ApoE mimetic improves pathology and memory in a model of Alzheimer’s disease. Brain Res. 1733, 146685 (2020).
Nelson, M. R. et al. The APOE-R136S mutation protects against APOE4-driven Tau pathology, neurodegeneration and neuroinflammation. Nat. Neurosci. 26, 2104–2121 (2023).
Chen, Y. et al. APOE3ch alters microglial response and suppresses Aβ-induced tau seeding and spread. Cell 187, 428–445.e20 (2023).
Marino, C. et al. APOE Christchurch-mimetic therapeutic antibody reduces APOE-mediated toxicity and tau phosphorylation. Alzheimers Dement. 20, 819–836 (2023).
Finkel, R. S. et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 377, 1723–1732 (2017).
Mendell, J. R. et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 377, 1713–1722 (2017).
Zhao, L. et al. Intracerebral adeno-associated virus gene delivery of apolipoprotein E2 markedly reduces brain amyloid pathology in Alzheimer’s disease mouse models. Neurobiol. Aging 44, 159–172 (2016).
Jackson, R. J. et al. APOE2 gene therapy reduces amyloid deposition and improves markers of neuroinflammation and neurodegeneration in a mouse model of Alzheimer disease. Mol. Ther. 32, 1373–1386 (2024).
Rosenberg, J. B. et al. AAVrh.10-mediated APOE2 central nervous system gene therapy for APOE4-associated Alzheimer’s disease. Hum. Gene Ther. Clin. Dev. 29, 24–47 (2018).
Ben Mkaddem, S., Benhamou, M. & Monteiro, R. C. Understanding Fc receptor involvement in inflammatory diseases: from mechanisms to new therapeutic tools. Front. Immunol. 10, 811 (2019).
Litvinchuk, A. et al. Apolipoprotein E4 reduction with antisense oligonucleotides decreases neurodegeneration in a tauopathy model. Ann. Neurol. 89, 952–966 (2021).
Laffitte, B. A. et al. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc. Natl Acad. Sci. USA 98, 507–512 (2001).
Boehm-Cagan, A. & Michaelson, D. M. Reversal of apoE4-driven brain pathology and behavioral deficits by bexarotene. J. Neurosci. 34, 7293–7301 (2014).
Cramer, P. E. et al. ApoE-directed therapeutics rapidly clear β-amyloid and reverse deficits in AD mouse models. Science 335, 1503–1506 (2012).
Ghosal, K. et al. A randomized controlled study to evaluate the effect of bexarotene on amyloid-β and apolipoprotein E metabolism in healthy subjects. Alzheimers Dement. (N. Y.) 2, 110–120 (2016).
Cummings, J. L. et al. Double-blind, placebo-controlled, proof-of-concept trial of bexarotene **n moderate Alzheimer’s disease. Alzheimers Res. Ther. 8, 4 (2016).
Boehm-Cagan, A. et al. ABCA1 agonist reverses the ApoE4-driven cognitive and brain pathologies. J. Alzheimers Dis. 54, 1219–1233 (2016).
Brodbeck, J. et al. Structure-dependent impairment of intracellular apolipoprotein E4 trafficking and its detrimental effects are rescued by small-molecule structure correctors. J. Biol. Chem. 286, 17217–17226 (2011).
Chen, H. K. et al. Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons. J. Biol. Chem. 287, 5253–5266 (2012).
Mahley, R. W. & Huang, Y. Small-molecule structure correctors target abnormal protein structure and function: the structure corrector rescue of apolipoprotein E4-associated neuropathology. J. Med. Chem. 55, 8997–9008 (2012).
Petros, A. M. et al. Fragment-based discovery of an apolipoprotein E4 (apoE4) stabilizer. J. Med. Chem. 62, 4120–4130 (2019).
Wang, C. et al. Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat. Med. 24, 647–657 (2018).
Husain, M. A., Laurent, B. & Plourde, M. APOE and Alzheimer’s disease: from lipid transport to physiopathology and therapeutics. Front. Neurosci. 15, 630502 (2021).
Raulin, A.-C. et al. ApoE in Alzheimer’s disease: pathophysiology and therapeutic strategies. Mol. Neurodegener. 17, 72 (2022).
Flowers, S. A., Grant, O. C., Woods, R. J. & Rebeck, G. W. O-glycosylation on cerebrospinal fluid and plasma apolipoprotein E differs in the lipid-binding domain. Glycobiology 30, 74–85 (2020).
Wahrle, S. E. et al. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J. Biol. Chem. 279, 40987–40993 (2004).
Sienski, G. et al. APOE4 disrupts intracellular lipid homeostasis in human iPSC-derived glia. Sci. Transl. Med. 13, eaaz4564 (2021).
Yeh, F. L., Wang, Y., Tom, I., Gonzalez, L. C. & Sheng, M. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron 91, 328–340 (2016).
Cooper, J. M. et al. Regulation of tau internalization, degradation, and seeding by LRP1 reveals multiple pathways for tau catabolism. J. Biol. Chem. 296, 100715 (2021).
Rauch, J. N. et al. LRP1 is a master regulator of tau uptake and spread. Nature 580, 381–385 (2020).
Holmes, B. B. et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl Acad. Sci. USA 110, E3138–E3147 (2013).
Reiman, E. M. et al. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proc. Natl Acad. Sci. USA 106, 6820–6825 (2009).
Sperling, R. A. et al. Association of factors with elevated amyloid burden in clinically normal older individuals. JAMA Neurol. 77, 735–745 (2020).
Jansen, W. J. et al. Prevalence estimates of amyloid abnormality across the Alzheimer disease clinical spectrum. JAMA Neurol. 79, 228–243 (2022).
Insel, P. S., Hansson, O. & Mattsson-Carlgren, N. Association between apolipoprotein E ε2 vs ε4, age, and β-amyloid in adults without cognitive impairment. JAMA Neurol. 78, 229–235 (2021).
Ramanan, V. K. et al. Association of apolipoprotein E ɛ4, educational level, and sex with tau deposition and tau-mediated metabolic dysfunction in older adults. JAMA Netw. Open 2, e1913909 (2019).
Young, C. B. et al. APOE effects on regional tau in preclinical Alzheimer’s disease. Mol. Neurodegener. 18, 1 (2023).
Therriault, J. et al. Association of apolipoprotein E ε4 with medial temporal tau independent of amyloid-β. JAMA Neurol. 77, 470–479 (2020).
Steward, A. et al. ApoE4 and connectivity-mediated spreading of tau pathology at lower amyloid levels. JAMA Neurol. 80, 1295–1306 (2023).
Ferrari-Souza, J. P. et al. APOEε4 potentiates amyloid β effects on longitudinal tau pathology. Nat. Aging 3, 1210–1218 (2023).
Morris, J. C. et al. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann. Neurol. 67, 122–131 (2010).
Maxwell, S. S. et al. Genetic associations with brain microbleeds: systematic review and meta-analyses. Neurology 77, 158–167 (2011).
Knol, M. J. et al. Association of common genetic variants with brain microbleeds: a genome-wide association study. Neurology 95, e3331–e3343 (2020).
Charidimou, A. et al. APOE and cortical superficial siderosis in CAA: meta-analysis and potential mechanisms. Neurology 93, e358–e371 (2019).
Auriel, E. et al. Validation of clinicoradiological criteria for the diagnosis of cerebral amyloid angiopathy-related inflammation. JAMA Neurol. 73, 197–202 (2016).
Theodorou, A. et al. Clinical, neuroimaging, and genetic markers in cerebral amyloid angiopathy-related inflammation: a systematic review and meta-analysis. Stroke 54, 178–188 (2023).
Reiman, E. M. et al. Declining brain activity in cognitively normal apolipoprotein E ε4 heterozygotes: a foundation for using positron emission tomography to efficiently test treatments to prevent Alzheimer’s disease. Proc. Natl Acad. Sci. USA 98, 3334–3339 (2001).
Reiman, E. M. et al. Correlations between apolipoprotein E ε4 gene dose and brain-imaging measurements of regional hypometabolism. Proc. Natl Acad. Sci. USA 102, 8299–8302 (2005).
Langbaum, J. B. S. et al. Hypometabolism in Alzheimer-affected brain regions in cognitively healthy Latino individuals carrying the apolipoprotein E ε4 allele. Arch. Neurol. 67, 462–468 (2010).
Knopman, D. S. et al. 18F-fluorodeoxyglucose positron emission tomography, aging, and apolipoprotein E genotype in cognitively normal persons. Neurobiol. Aging 35, 2096–2106 (2014).
Malek-Ahmadi, M. et al. Plasma NfL is associated with the APOE ε4 allele, brain imaging measurements of neurodegeneration, and lower recall memory scores in cognitively unimpaired late-middle-aged and older adults. Alzheimers Res. Ther. 15, 74 (2023).
Strom, A. et al. Cortical hypometabolism reflects local atrophy and tau pathology in symptomatic Alzheimer’s disease. Brain 145, 713–728 (2022).
Salvadó, G. et al. The protective gene dose effect of the APOE ε2 allele on gray matter volume in cognitively unimpaired individuals. Alzheimers Dement. 18, 1383–1395 (2022).
Butt, O. H. et al. Cognitively normal APOE ε4 carriers have specific elevation of CSF SNAP-25. Neurobiol. Aging 102, 64–72 (2021).
Sun, X. et al. APOE ε4 carriers may undergo synaptic damage conferring risk of Alzheimer’s disease. Alzheimers Dement. 12, 1159–1166 (2016).
Ferrari-Souza, J. P. et al. APOEε4 associates with microglial activation independently of Aβ plaques and tau tangles. Sci. Adv. 9, eade1474 (2023).
Benedet, A. L. et al. Differences between plasma and cerebrospinal fluid glial fibrillary acidic protein levels across the Alzheimer disease continuum. JAMA Neurol. 78, 1471–1483 (2021).
Spotorno, N. et al. Astrocytic function is associated with both amyloid-β and tau pathology in non-demented APOE ε4 carriers. Brain Commun. 4, fcac135 (2022).
Operto, G. et al. Interactive effect of age and APOE-ε4 allele load on white matter myelin content in cognitively normal middle-aged subjects. NeuroImage Clin. 24, 101983 (2019).
Triebswetter, C. et al. Differential associations between apolipoprotein E alleles and cerebral myelin content in normative aging. NeuroImage 251, 118988 (2022).
Janelidze, S. et al. Increased blood–brain barrier permeability is associated with dementia and diabetes but not amyloid pathology or APOE genotype. Neurobiol. Aging 51, 104–112 (2017).
Cicognola, C. et al. Associations of CSF PDGFRβ with aging, blood–brain barrier damage, neuroinflammation, and Alzheimer disease pathologic changes. Neurology 101, e30–e39 (2023).
Mahley, R. W. & Rall, S. C. Is ε4 the ancestral human apoE allele? Neurobiol. Aging 20, 429–430 (1999).
Seixas, S., Trovoada, M. J. & Rocha, J. Haplotype analysis of the apolipoprotein E and apolipoprotein C1 loci in Portugal and São Tomé e Príncipe (Gulf of Guinea): linkage disequilibrium evidence that APOE*4 is the ancestral APOE allele. Hum. Biol. 71, 1001–1008 (1999).
Fullerton, S. M. et al. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am. J. Hum. Genet. 67, 881–900 (2000).
Smith, C. J. & Ashford, J. W. Apolipoprotein ɛ4-associated protection against pediatric enteric infections is a survival advantage in pre-industrial populations. J. Alzheimers Dis. 93, 907–918 (2023).
Trumble, B. C. et al. Apolipoprotein-ε4 is associated with higher fecundity in a natural fertility population. Sci. Adv. 9, eade9797 (2023).
Ostendorf, B. N. et al. Common germline variants of the human APOE gene modulate melanoma progression and survival. Nat. Med. 26, 1048–1053 (2020).
Zokaei, N. et al. Short-term memory advantage for brief durations in human APOE ε4 carriers. Sci. Rep. 10, 9503 (2020).
Lancaster, C., Forster, S., Tabet, N. & Rusted, J. Putting attention in the spotlight: the influence of APOE genotype on visual search in mid adulthood. Behav. Brain Res. 334, 97–104 (2017).
Lancaster, C., Tabet, N. & Rusted, J. The APOE paradox: do attentional control differences in mid-adulthood reflect risk of late-life cognitive decline. Neurobiol. Aging 48, 114–121 (2016).
Lancaster, C., Tabet, N. & Rusted, J. The elusive nature of APOE ε4 in mid-adulthood: understanding the cognitive profile. J. Int. Neuropsychol. Soc. 23, 239–253 (2017).
Lu, K. et al. Dissociable effects of APOE-ε4 and β-amyloid pathology on visual working memory. Nat. Aging 1, 1002–1009 (2021).
Zink, N., Bensmann, W., Arning, L., Beste, C. & Stock, A.-K. Apolipoprotein ε4 is associated with better cognitive control allocation in healthy young adults. NeuroImage 185, 274–285 (2019).
Sullivan, P. M. et al. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J. Biol. Chem. 272, 17972–17980 (1997).
Sullivan, P. M., Mezdour, H., Quarfordt, S. H. & Maeda, N. Type III hyperlipoproteinemia and spontaneous atherosclerosis in mice resulting from gene replacement of mouse Apoe with human Apoe*2. J. Clin. Invest. 102, 130–135 (1998).
Raffai, R. L., Dong, L. M., Farese, R. V. & Weisgraber, K. H. Introduction of human apolipoprotein E4 ‘domain interaction’ into mouse apolipoprotein E. Proc. Natl Acad. Sci. USA 98, 11587–11591 (2001).
Foley, K. E. et al. The APOE ε3/ε4 genotype drives distinct gene signatures in the cortex of young mice. Front. Aging Neurosci. 14, 838436 (2022).
Piedrahita, J. A., Zhang, S. H., Hagaman, J. R., Oliver, P. M. & Maeda, N. Generation of mice carrying a mutant apolipoprotein E gene inactivated by gene targeting in embryonic stem cells. Proc. Natl Acad. Sci. USA 89, 4471–4475 (1992).
Raber, J. et al. Isoform-specific effects of human apolipoprotein E on brain function revealed in ApoE knockout mice: increased susceptibility of females. Proc. Natl Acad. Sci. USA 95, 10914–10919 (1998).
Sun, Y. et al. Glial fibrillary acidic protein-apolipoprotein E (apoE) transgenic mice: astrocyte-specific expression and differing biological effects of astrocyte-secreted apoE3 and apoE4 lipoproteins. J. Neurosci. 18, 3261–3272 (1998).
Golden, L. R. & Johnson, L. A. APOE allele switching in a novel transgenic mouse model as a therapeutic approach for Alzheimer’s disease. Alzheimers Dement. 18, e060213 (2022).
Acknowledgements
NIH/NIA (2RF1AG047644 to R.J.J. and B.T.H., 1RF1AG073236 to R.J.J., R56AG080525 to R.J.J. and B.T.H., 5U01NS111671 to B.T.H., 5K08AG064039 to A.S.-P. and P30AG062421 to A.S.-P. and B.T.H.), The Karen Toffler Charitable Trust (to A.S.-P. and B.T.H.), The Harrison Gardner Jr Innovation Award (to A.S.-P.) and The JPB Foundation (to B.T.H.).
Author information
Authors and Affiliations
Contributions
R.J.J. and A.S.-P. reviewed the literature and wrote the manuscript draft. B.T.H. reviewed and edited the draft.
Corresponding authors
Ethics declarations
Competing interests
R.J.J. declares no competing interest. B.T.H. serves on the SAB of Latus and of Dewpoint and has a family member who is employed by Novartis. A.S.-P. has signed a material transfer agreement with Ionis Pharmaceuticals, Inc.
Peer review
Peer review information
Nature Reviews Neurology thanks Takahisa Kanekiyo, Shahram Oveisgharan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Adeno-associated viral vectors
-
Small non-pathogenic viruses that can infect cells and deliver a small single-stranded DNA cargo of <5 kb. This DNA is then transcribed and translated by the target cell generating the protein of interest.
- Antisense oligonucleotides
-
(ASOs). ASOs are short RNA transcripts that are synthesized to be complementary to the sequence of a specific RNA target with the goals of preventing its translation into the protein and promoting its degradation. ASOs are often chemically modified to increase stability (resistance to degradation by RNAse enzymes) and enhance cellular uptake.
- ATP-binding cassette (ABC) transporters such as ABCA1 and ABCG
-
Transmembrane proteins that transport cholesterol and phospholipids out of the cell to lipid-poor apolipoproteins such as APOE.
- Cell-autonomous and non-autonomous
-
Cell-autonomous effects are those that a perturbed cell exerts on itself or other cells of the same type. Cell-non-autonomous effects are those that a perturbed cell type (for example, astrocytes) exerts on other cell types (for example, microglia), either directly or via its secretome.
- Exposome
-
Set of non-genetic risk factors that can impact the risk of develo** certain disease (for example, cancer or Alzheimer disease) of an individual, including cumulative lifetime environmental exposures and lifestyle habits.
- Global genetic ancestry
-
Genetic variability across the genome that determines the race and ethnicity of an individual based on the relative proportions of various population ancestries (for example, European, African, Amerindian or East Asian), not always coincident with self-reported categories.
- Human-inducible pluripotent stem cells
-
(hiPSCs). Cells derived from skin or blood cells after reprogramming them back to a pluripotent embryonic-like state, which can be then differentiated to recapitulate any main brain cell type, although often embryonic or fetal in nature. Isogenic versions that are genetically identical except for the gene of interest (for example, APOE) can be generated with CRISPR–Cas9 technology.
- Local genetic ancestry
-
Genetic variability surrounding a specific locus in the genome of an individual, which can include zero, one or two copies of an allele from each ancestral population, thereby affecting the expression of a gene of interest (for example, APOE).
- Polygenic risk score
-
(PRS). An estimate of the genetic relative risk of an individual to develop a certain disease, calculated by applying the summary statistics from meta-analysis of genome-wide association studies involving thousands of cases and controls to the genetic variants of that particular individual.
- Triggering receptor expressed on myeloid cells 2
-
(TREM2). A receptor expressed on the cell surface of immune cells, including microglia, that activates phagocytosis in response to extracellular stress signals (for example, Aβ) through the TYROBP–DAP12 signalling pathway.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jackson, R.J., Hyman, B.T. & Serrano-Pozo, A. Multifaceted roles of APOE in Alzheimer disease. Nat Rev Neurol (2024). https://doi.org/10.1038/s41582-024-00988-2
Accepted:
Published:
DOI: https://doi.org/10.1038/s41582-024-00988-2
- Springer Nature Limited