Abstract
Light-based 3D bioprinting is now employed widely to fabricate geometrically complex constructs for various biomedical applications. However, the inherent light scattering defect creates significant challenges in patterning dilute hydrogels to form high-fidelity structures with fine-scale features. Herein, we introduce a photoinhibiting approach that can effectively suppress the light scattering effect via a mechanism of simultaneous photoabsorption and free-radical reaction. This biocompatible approach significantly improves the printing resolution (~1.2 - ~2.1 pixels depending on swelling) and shape fidelity (geometric error less than 5%), while minimising the costly trial-and-error procedures. The capability in patterning 3D complex constructs using different hydrogels is demonstrated by manufacturing various scaffolds featuring intricate multi-sized channels and thin-walled networks. Importantly, cellularised gyroid scaffolds (HepG2) are fabricated successfully, exhibiting high cell proliferation and functionality. The strategy established in this study promotes the printability and operability of light-based 3D bioprinting systems, allowing numerous new applications for tissue engineering.
Similar content being viewed by others
Introduction
Three-dimensional (3D) bioprinting technology, which allows cells and biomaterials to be patterned precisely, is a promising tool for fabricating highly sophisticated constructs for biomedical applications, such as tissue engineering, drug testing and surgical implants1,2,3,4,5,6. Among various 3D bioprinting modalities, digital light processing-based (DLP) printing has recently gained immense popularity due to its fine resolution (a few micrometres), rapid printing speed (seconds to minutes printing time) and predominant spatiotemporal controllability1. This technology converts liquid photocurable prepolymer into structured objects in a layer-wise fashion7,8 or via volumetric projection9 and has advanced bioprinting technology to create more elaborate structures, multi-vascular networks10, cell-laden scaffolds with multiscale channels11 and organ-on-a-chip systems12 for instance. In addition, DLP printers with high optical resolution (typically 1–50 μm) offer the possibility for fabricating 3D hydrogel constructs embedded with fine-scale features necessary to adequately recapitulate the complexities within the native tissue microenvironment and thus achieve appropriate biological behaviours in vitro13,14,15,28 and Ponceau 4R29, anionic azo dye30 and 2-hydroxy-4-methoxy benzophenone-5-sulfonic acid (HMBS)31 that have enabled the reproduction of convoluted constructs embedded with multi-channels and vascular network23,29,32. However, the performance of the standard photoabsorbers is limited, deteriorating the capacity of the DLP printers they should possess1. Such an effect is more pronounced for platforms with a higher optical resolution33, particularly when cells are encapsulated25, creating significant challenges for patterning 3D cellularised constructs with fine-scale features (such as intricate micro-sized channels and thin-walled networks) that are of great importance to nutrient transportation and oxygen permeation. Notably, the existing photoabsorbing-based approach requires intensive trial-and-error efforts to optimise the printing parameters (e.g., light intensity) and bioink formulation (e.g., the concentration of each component) to achieve the desired pattern fidelity for a specific structure, tremendously hindering the practical use of DLP-based bioprinting technologies17,34. Such a trial-and-error approach is not only time-consuming and costly but also challenging to obtain the most optimal printing accuracy. Furthermore, previous studies have revealed that bioink with low-concentration hydrogels exhibits better cytocompatibility35; however, such a dilute nature imposes extra difficulties on the physical printing process. A fundamental issue of hydrogel photopatterning is how to simultaneously improve the printability for building complex constructs with high fidelity, simplify the operability for easy practical use and maintain appropriate biocompatibility for facilitating cell activities, a significant challenge in light-based 3D bioprinting technology.
Besides the photoinhibiting strategy, You et al. proposed a flashing photopolymerisation technique to suppress the scattering effect by minimising the light exposure time19. However, the cell-induced scattering effects cannot be resolved by using this method, making it inapplicable for fabricating cell-laden constructs with fine features. Recently, Guan et al. introduced a machine-learning approach to improve pattern fidelity by compensating for the scattering-induced geometric deviation when generating digital masks25,36. The intra-layer printing resolution can be enhanced using this method, yet performance for multi-layer fabrication of complex structures was not demonstrated. Therefore, a new biocompatible strategy that can effectively eliminate the light scattering effect is urgently required for light-based bioprinting.
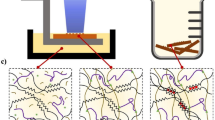
Ideally, if the free radicals generated by light scattering can be removed or lowered below a threshold (e.g., through chemical reactions), the polymerisation of hydrogel prepolymers beyond designated regions can be prohibited. This paper presents a photoinhibiting approach that promotes the fabrication resolution and pattern fidelity of DLP bioprinting by addressing the light scattering problem, as well as minimising the trial-and-error optimisation of printing parameters. Unlike typical photoabsorbers, the photoinhibiting additive newly developed in this study can control the fundamental polymerisation process via simultaneous photoabsorption and free-radical reaction. The photoabsorption that functions as conventional photoabsorbers can prevent over-curing layers by absorbing excess light, thus improving the vertical printing resolution. The free-radical reaction can rapidly consume the free radicals generated by light scattering so that hydrogel monomers are starved at free radicals to form photo-crosslinked hydrogels in scattered regions, thereby impeding unwanted solidification. With this method, the printing resolution and pattern fidelity are significantly enhanced, and various complex 3D structures with multiscale features (particularly 3D cellularised scaffolds) can be fabricated all at once without re-tuning the printing parameters.
Firstly, based on the mechanism above, we synthesised a new photoinhibiting additive (curcumin-Na (Cur-Na)) that is water-soluble and cytocompatible. Secondly, PEG-GelMA bioinks added with various photoinhibiting additives were prepared for comparison, and the physico-chemical characterisations were performed to assess their printability and mechanical properties. The underlying mechanism of how Cur-Na mitigates the scattering effects was then elucidated by examining the polymerisation process theoretically and experimentally. Thirdly, the printing resolution and accuracy were investigated quantitatively for the bioinks to verify Cur-Na’s effectiveness for resolving the light scattering defect. Various architecturally-complex constructs were successfully fabricated using the Cur-Na bioink without re-optimising the printing parameters, demonstrating superb capability in patterning 3D functional cellularised scaffolds with fine-scale features (e.g., gyroid). Finally, the cytocompatibility of the Cur-Na bioink was confirmed through in vitro culturing experiments.
Results and discussion
Characterisation of Cur-Na
To obtain a water-soluble reactive additive, we synthesised Cur-Na by chemically modifying curcumin using sodium bicarbonate, where the H+ of phenolic hydroxyl of curcumin molecular was substituted by Na+ (red ellipses in Fig. 1a). Meanwhile, the carbonyl bond in curcumin underwent keto-enol tautomerism due to the hydroxyl group in methanol, forming a new C=C double bond (blue ellipses in Fig. 1a). Such modifications were confirmed by proton nuclear magnetic resonance spectroscopy (1H-NMR). As shown in Fig. 1b, the characteristic resonance of the phenolic hydroxyl group (δ = 9.65 ppm) has disappeared by adding sodium to curcumin, indicating the substitution of H+. The formation of the C=C double bond was evidenced by the changes in the alkene signal (δ = 5.93 ppm), where a new signal peak representing the trisubstituted alkene was developed (Fig. 1b). The full spectrum of 1H-NMR and 13C-NMR are available in Supplementary Fig. 1 and Fig. 2. The existence of Na was determined by the 23Na-NMR spectrum (see Supplementary Fig. 3). The molecular weight measured by high-resolution mass spectrometry (HRMS) is 434, identical to that of the structural formula (see Supplementary Fig. 4 and Supplementary Table 1). The synthetic Cur-Na is a multifunctional additive that can be incorporated into bioink to improve printing quality. To verify its suitability for light-based 3D bioprinting, the light absorbance and water solubility were initially assessed experimentally. As shown in the absorbance spectra (Supplementary Fig. 5), Cur-Na demonstrated excellent light attenuation capability due to the absorbance peak (~425 nm) near 405 nm, a wavelength commonly used in light-based 3D bioprinting owing to its low detrimental effects on cells compared with UV light (e.g., 365 nm)37,9,10,17,30. It is also evident that the unresolved region is enlarged when light energy increases. When adding 1 mM tartrazine to the bioink, the resolution sensitivity on exposure intensity is still apparent despite the unresolved fraction for all energy doses being lowered. In the case of Cur-Na, the resolution is improved significantly, as indicated by the shrinking unresolved circles in the lower panel of Fig. 4c. The sensitivity to light intensity is minor, even though it is visually noticeable in the figure. To quantify the resolution difference, the relation between the unresolved fraction and relative energy is plotted in Fig. 4d. It can be seen that the resolution of the pure PEG-GelMA bioink is highly dependent on energy dosage, and the unresolved fraction is amplified from ~17% to as large as ~33% when the exposure intensity is tripled. A similar trend was observed for tartrazine, with reduced values that are ~12% and ~21% for Er = 1 and Er = 3, respectively. Unlike tartrazine, the unresolved fraction of Cur-Na remains at an almost constant value irrelevant to the exposure intensity, being as low as ~12%, suggesting that a fixed concentration of Cur-Na can deal with scattering with different intensities while preserving higher printing resolution. Figure 4e depicts the printing window that characterises the extent of exposure energy within which the printing resolution is maintained, serving as a quantitative guide for selecting the concentration of Cur-Na. For other photoabsorbers (e.g., Ponceau 4 R), a trend similar to tartrazine was observed due to their comparable absorbance peaks, being ~405 nm and ~500 nm for tartrazine and Ponceau 4R, respectively (see Supplementary Fig. 8).
When cells are encapsulated (see the fluorescent staining image in Fig. 4b), the scattering effect of the pure PEG-GelMA bioink is more pronounced, further increasing the unresolved areas and thus reducing the printing resolution (Fig. 4f and Supplementary Fig. 9). For tartrazine, when the exposure intensity that substantially governs the degree of scattering increases, the resolution is lowered due to enhanced scattering (Fig. 4f). A higher concentration of tartrazine might be required to compensate for the increased over-curing. However, it is at the expense of sacrificing the structure’s stiffness and printing time because there is a trade-off between printing resolution, printing efficiency and mechanical performance, mediated by the concentration of photoabsorber1,17,20. Purely increasing the amount of tartrazine or Ponceau 4R cannot improve the printing quality (see Supplementary Fig. 8c, d). Comparing the cell-free (Fig. 4d) and cell-laden (Fig. 4f) structures printed with Cur-Na, the influence of the cell-enhanced scattering on printing resolution is ignorable due to the scattering suppression mechanism of Cur-Na described previously. The results above reveal that Cur-Na can improve printing resolution better than a purely photoabsorbing-based method and that a fixed low concentration of Cur-Na is sufficient to suppress the scattering effect of various strengths, therefore simplifying the operational process. For a purely photoabsorbing-based method, the concentration of a photoabsorber (e.g., tartrazine) needs to be adjusted to match a unique scattering characteristic to achieve a better printing resolution17. However, the scattering characteristic varies dramatically along with changes in light intensity, formulation of bioink, exposure time, shape of structures and cell density34.
Matching the printing resolution with the optical resolution of printers is one of the greatest challenges for DLP printing because of the light scattering issue and the dilute nature of the hydrogels (swelling and low viscosity)32. The sensitivity of printing resolution to the concentration of hydrogels and various photoinhibitors was investigated to understand how light scattering and swelling impact the printing resolution (see Supplementary Fig. 10). The PEG-GelMA/Cur-Na hydrogel used in this study demonstrated a printing resolution of ~2.1 pixels, while a resolution (~1.2 pixels) of proximity to the printer’s optical resolution was achieved when the concentration of PEGDA was increased to 30% due to weakened swelling. The full printing capacity of DLP printers (1 pixel) is thus achievable using the method proposed in this study once the swelling of printing ink is confined entirely. For 30% PEGDA hydrogels, the resolution dropped from ~1.2 to ~2.7 pixels when replacing Cur-Na with standard photoabsorbers, although extensive parameter tuning was conducted. Such a change is due mainly to the scattering effect because the swelling ratio is almost identical, indicating that Cur-Na can suppress light scattering more effectively.
We then investigated how the scattering effect during multi-layer fabrication can be inhibited by Cur-Na, thus improving the printing quality of 3D structures. Biofabricated hollow structures are of particular importance for biomedical applications because they are essential for guiding tissue regeneration, forming vascular networks and accommodating cell attachment and growth1,11,5g). Figure 5h compares the designed diameters and those measured, where the diameters of blocked channels were assumed to be zero. It can be seen that the dimensions of the channels printed using Cur-Na match very well with those designed, and the average error is less than ~5%, indicating high printing accuracy. For tartrazine, the measured diameters are narrowed by ~50 μm owing to the over-curing compared with those designed, although we carefully optimised the concentration of tartrazine. Huh et al. investigated the impact of the concentration of the most used photoabsorbers on the printing accuracy of DLP17. They demonstrated that a fixed concentration of photoabsorbers cannot effectively inhibit the scattering effect if the diameter of the channel is too small, leading to over-curing and blockage. In contrast, light is over-blocked when the diameter increases, resulting in a larger printed diameter than that designed. There is a transition from over-curing to insufficient polymerisation owing to the dynamic changes in scattering intensity. Consequently, the concentration of photoabsorbers should be matched precisely with the scattering characteristic of a structure to minimise printing errors, creating significant challenges for practical use. Unlike photoabsorbers, Cur-Na is self-adaptive to the dynamic changes of scattering characteristics, allowing various high-fidelity 3D constructs to be fabricated without re-optimising printing parameters, as further highlighted in the next section.
Performance of printing 3D structures
To further demonstrate Cur-Na’s printability of complex 3D structures, convoluted scaffolds, including spinal cord scaffold, blood vessels and gyroid scaffold, were fabricated using the PEG-GelMA/Cur-Na bioink (Fig. 6). Neural scaffolds featuring parallel-aligned channels were usually adopted for biomimicking living neural constructs for spinal cord injury repair in previous studies23,44,45,46. A newly designed spinal cord scaffold was fabricated successfully using the Cur-Na bioink, showing irregular channels and thin-walled networks that can better mimic the internal architecture of the spinal cord (Fig. 6a and Supplementary Fig. 12a). The thickness of the thin-walled networks ranges from approximately 80 to 100 μm essential for effective exchange of metabolites and oxygen permeation47. Vascular networks with sizes of ~150–200 μm are necessary to establish adequate vascularisation for long-term cell activities; yet, it remains a considerable challenge for engineering tissue with vascular networks proximity to the size of capillary34,48. Based on our previously designed 3D vessel model for skin tissue regeneration49,50, a vascular network with multiple branches and perfusable channels (800–200 μm) was fabricated using the same bioink formulation (Fig. 6b), demonstrating high capability in generating vessels with vast length scales. The vascular wall (~100 μm) was well preserved, which differs from the previous DLP-printed hydrogel vascular networks10,17,32, where the channels were embedded within a bulk of hydrogel. The perfusion of the vascular network was confirmed via injection with a coloured dye solution (Fig. 6b and Supplementary Movie 1). Gyroid scaffolds made of quadruple junction points have shown promise in tissue scaffolding due to their unique topological features (e.g., curved surface, high porosity and interconnectivity) that yield optimal nutrient and waste diffusion for cell activities29,50. However, the architectural richness gives rise to the thin-walled structures that are difficult to manufacture. Most applications of 3D-printed gyroid scaffolds focus on hard tissue repair, and the printing materials (e.g., poly(ε-caprolactone) and hydroxyapatite) usually exhibit high stiffness51,52. Functional cellularised gyroid scaffolds engineered with hydrogels were, however, rarely reported. Huh et al. DLP printed a cell-laden gyroid scaffold at a scale of centimetres using the photoabsorbing-based method17. However, the cell viability and functionality were not demonstrated. In this study, gyroid scaffolds were successfully fabricated using the Cur-Na bioink, allowing the formation of ordered pores with a size of 150 μm (Fig. 6c). To demonstrate the performance for cell functionality, gyroid scaffolds encapsulated with HepG2 cells (1 × 107 mL−1) that are susceptive to light scattering were also fabricated successfully. High cell proliferation was observed after culturing for 14 days (Fig. 6d). Importantly, sphere formation was observed on day 7, and the protein expression of albumin increased rapidly after (see Supplementary Fig. 13), indicating enhanced hepatic functions. The cellularised gyroid scaffolds engineered with hydrogel offer numerous new applications for tissue engineering.
a Spinal cord scaffold designed based on micro-CT scanning data. It features irregular channels and thin-walled networks that mimic the internal architecture of the spinal cord. The scaffolds were fabricated successfully using two types of hydrogels: PEG-GelMA (10%) and silk fibroin modified by glycidyl methacrylate (Sil-MA (15%)). b Vascular network with multiple branches, perfusable channels (200–800 μm) and thin walls (~100 μm). The perfusion was performed via injection with a coloured dye solution. c Gyroid scaffold with unique topological features (curved surface, high porosity and interconnectivity). d Printed gyroid scaffold encapsulated with HepG2 cells and cultured for 14 days (confocal images), showing high cell proliferation. Images a–d are representatives of n = 3 independent experiments.
To demonstrate the feasibility of Cur-Na for other hydrogels, the spinal cord scaffold was fabricated using the Sil-MA7 bioink that has a printable window of 15–30% in concentration. Sil-MA bioink is opaque (strong light scattering) and the printed parts usually possess low compressive properties, thus more appropriate for exploring the performance boundary of Cur-Na. The lowest value of the printing window (15%) was employed in this study, although a concentration of 30% Sil-MA mixed with standard photoabsorber was used by Kim et al. to demonstrate its capability for patterning 3D complex structures7. Surprisingly, the scaffold was fabricated successfully using Cur-Na, precisely reproducing the designed fine-scale features (see Supplementary Fig. 12b). With standard photoabsorbers, the printing outcomes were, however, not ideal due mainly to intensive light scattering, demonstrating twisted thin-walled networks and blocked channels. Notably, the printed scaffold exhibited a low Young’s modulus (8–12 kPa depicted in Supplementary Fig. 12c, d) that is ~9× softer than the PEG-GelMA hydrogel used in this study, expanding the potential for constructing soft tissues such as liver and heart organs53.
Cytocompatibility
To evaluate the cytocompatibility of Cur-Na for other cells, live/dead and CCK-8 assays for PC-12 cells were carried out for the three groups of bioink developed previously (Fig. 7a, b). It can be observed that the PEG-GelMA/Cur-Na hydrogel exhibited the uppermost cell viability, and the cell proliferation was approximately tripled after 14 days in culture, even higher than that of the pure PEG-GelMA bioink. The superior cytocompatibility is likely attributed to that Cur-Na is derived from curcumin, a natural phenolic antioxidant widely used for antioxidant, anti-inflammatory, antimicrobial, and so on54. Interestingly, the cell proliferation of the PEG-GelMA/Tartrazine hydrogel was not observed, even though tartrazine has been recognised as biocompatible and low-toxicity10.
a Live and dead assay stained images showing cell viability of encapsulated PC-12 cells for 14 days (live cells in green and dead cells in red). Scale bars indicate 100 μm. b CCK-8 assay; PC-12 cell proliferation in the cell-laden samples for 14 days. The PEG-GelMA/Cur-Na hydrogel exhibited the best cell proliferation. Each assay was conducted in triplicate. n = 3 independent samples. c Fluorescent images of the channelled scaffold (a diameter of 200 μm) fabricated using the PEG-GelMA/Cur-Na hydrogel, showing homogeneous cell distribution. The last image displays the cell growth along the channels. Scale bars indicate 500 μm. d Confocal images of the middle channel of the spinal cord scaffold on days 1, 7, and 14. Scale bars indicate 500 μm. e Fluorescence images showing differentiation and neurite formation of PC-12 cells (day 7) on PEG-GelMA/Cur-Na scaffold where the cells were seeded on the surface of the scaffold: Beta3-Tubulin (Tuj-1) and 4’,6-diaminyl-2-phenylindole (DAPI). White scale bars indicate 100 μm. Neurite formation was observed on day 7. Images (c–e) are representatives of n = 3 independent experiments. Data are presented as mean values ± standard deviation. The significant difference is determined by two-tailed t test, *P < 0.05, **P < 0.01, ***P < 0.001 and P > 0.05 (no significant difference).
To further demonstrate the suitability for tissue engineering applications, we fabricated a cell-laden scaffold featuring multiple channels (200 μm) using the Cur-Na bioink (Fig. 7c), followed by in vitro culture experiments to assess the cell growth behaviours. During the printing process, each layer with a thickness of 100 μm was printed within a relatively short period (10 s), which can effectively prevent the settling of cells. It can be seen that the cell distribution is homogeneous within the scaffold (Fig. 7c), which is key to enhancing cell communication and avoiding the cell aggregation effect. Microchannels with a diameter of 200 μm were patterned at a high concentration of cells (1–2 × 107 mL−1), further highlighting Cur-Na’s printing performance for generating cell-laden 3D constructs. After 14 days in culture, the cells grew well along the wall of the channel (Fig. 7c, d), suggesting that the microchannels are capable of guiding cell growth, which can facilitate central nervous system regeneration23,45 and vascular network formation49. The structure remained stable after 14-day culturing; the size of channels was however amplified compared to that of day 1. It is likely due to the degradation phenomenon that can occur within hydrogel materials8. To verify whether Cur-Na and the generated hydrogel influence the differentiation behaviours of PC-12 cells, two differentiation tests were conducted by culturing PC-12 cells in the differentiation medium containing 1 mM Cur-Na (Supplementary Fig. 14) and by seeding the cells on the surface of the generated 3D channelled scaffold (Fig. 7e). After 7-day culturing, both differentiation and neurite formation were observed, demonstrating the potential for cell functionality studies.
In summary, we introduced an innovative photoinhibiting approach that can effectively suppress the inherent light scattering effect of light-based 3D printing platforms via a mechanism of simultaneous photoabsorption and free-radical reaction. We then synthesised a photoinhibiting additive (Cur-Na) based on such a mechanism and showcased its advantages by do** bioink with this additive, including (1) promoting the printing resolution and fidelity significantly. The full capacity of DLP printers (1 pixel) is achievable once the swelling of printing ink is confined; (2) simplifying the operating process by eliminating the re-optimisation procedures, which addresses the trial-and-error issue and has important practical and economic meaning; (3) superb performance in fabricating complex constructs featuring intricate micro-sized channels and thin-walled networks that are of great importance for nutrient transportation and oxygen permeation; (4) cell-laden gyroid scaffolds (HepG2 cells) that are susceptive to light scattering are fabricated successfully, exhibiting high cell proliferation and hepatic functions after 14-day culturing. (5) This mechanism is generic and applicable for other printing inks (e.g., Sil-MA) as long as the free-radical chain-growth polymerisation governs the printing process. This methodology offers a straightforward way to impart multiscale features into tissue constructs, which is vital for better mimicking the native tissue microenvironment. The strategy established in this study promotes the printability and operability of 3D bioprinting in patterning complex constructs with high fidelity, allowing for more advanced tissue engineering and regenerative medicine.
Methods
Curcumin-Na synthesis
Curcumin-Na was synthesised by chemically modifying curcumin using sodium bicarbonate. Specifically, 368 mg (1 mmol) curcumin (purity ≥98%, Aladdin) was dissolved in methyl alcohol (4 mL, purity ≥99.9%, Sigma-Aldrich) and deionised water (4 mL), and stirred until complete dissolution. Overall, 294 mg (3.5 mmol) NaHCO3 (purity ≥99%, Sigma-Aldrich) was then added to the solution to react with curcumin at ambient temperature. The mixture was dried by rotary evaporation and dissolved in 3 mL of anhydrous dichloromethane (purity ≥97%, Sigma-Aldrich). Filtering was performed subsequently to remove the insoluble matter. Finally, the solution was freeze-dried to obtain Cur-Na powder, and the yield was 82.9%. The Cur-Na powder was stored at −20 °C for further use. Note that methyl alcohol is toxic, and the synthetic process must be carried out under a ventilated condition.
Characterisation (HRMS/NMR)
To determine the molecular weight of Cur-Na, the mass spectrum of Cur-Na was obtained by using high resolution mass spectrometer (HRMS) with a scanning range of 100–1000 m z−1. Cur-Na was manipulated and monitored under the electrospray ionisation (ESI) positive mode (Bruker Daltonics, Germany). The constituents of Cur-Na were identified by nuclear magnetic resonance (NMR) and the data were analysed using MestReNova14 software. 1H-NMR (400 MHz, DMSO-d6), 13C-NMR (101 MHz, DMSO-d6) and 23Na-NMR (159 MHz, DMSO-d6) spectra characterisation were performed via an NMR spectrometer (Bruker Avance III, Switzerland) using DMSO-d6 (purity ≥99.9%, Sigma-Aldrich) as the solvent. The samples were 0.7 mL DMSO-d6/6 mg Cur-Na, 0.9 mL DMSO-d6/25 mg Cur-Na and 0.9 mL DMSO-d6/20 mg Cur-Na for 1H-NMR, 13C-NMR and 23Na-NMR, respectively. The detailed peak list of NMR spectra and HMRS data are available in Supplementary Figs. 1–4 and Supplementary Table 1, respectively.
Polymer and photoinitiator synthesis
Poly (ethylene glycol) diacrylate (6 kDa PEGDA) was synthesised following the protocol of Miller et al.55. Specifically, dry poly (ethylene glycol) (PEG; MW 6000, 81260, Sigma-Aldrich) was dissolved in anhydrous dichloromethane (purity ≥97%, Sigma-Aldrich). Triethylamine (2 molar over-dose to PEG, purity ≥99.5%, Sigma-Aldrich) and acryloyl chloride (4 molar over-dose to PEG, purity ≥97%, Sigma-Aldrich) were added into the mixture. The process was reacted overnight under argon gas. The yield was ~80%. The synthesised PEGDA was stored at −20 °C for further use. The synthetic process must be carried out under ventilated conditions.
Gelatin methacryloyl (GelMA) was synthesised following the protocol of Shirahama et al.56 with slight modifications. Specifically, 10 g gelatin (G1890-100G, Sigma-Aldrich) was added to PBS by stirring and the mixture was heated to 60 °C to obtain a gelatin solution. In total, 1.25 mL methacrylic anhydride (purity ≥ 94%, Macklin) was slowly added to the gelatin solution with continuous stirring. The mixture was reacted at 60 °C for 3 h under a pH from 8.0 to 9.0; the resulting products were dialysed in ddH2O for 3 days. The GelMA solution was frozen at −80 °C and lyophilised for 3 days. The yield was about 80%. The GelMA was stored at −20 °C for further use. The synthetic process must be carried out under ventilated conditions.
The photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) was synthesised following the method by Fairbanks et al.57. Specifically, dimethyl phenylphosphonite (0.186 mL, 1.17 mmol, purity ≥97%, Sigma-Aldrich) was added to an equivalent mole of 2,4,6-trimethylbenzoyl chloride (purity ≥97%, Sigma-Aldrich) and the mixture was then stirred overnight at room temperature under argon gas. The resultant products were mixed with excess lithium bromide (0.406 g, 4.68 mmol, purity ≥99%, Sigma-Aldrich) and 2-butanone (10 mL, purity ≥99%, Sigma-Aldrich). The mixture was subsequently heated to 50 °C for 20 min to ensure a complete reaction. Afterwards, the reaction products were rested for at least 4 h and cooled down to room temperature, followed by filtering the solution. The unreacted lithium bromide in the filtrate was then removed by adding the 2-butanone solvent, and the excess solvent was removed by rotary evaporation. The yield was about 80%. The LAP was stored at −20 °C for further use. The synthetic process must be carried out under ventilated conditions.
PEG-GelMA bioink composed of 6 wt% PEGDA (6 kDa), 4 wt% GelMA and 17 mM LAP was developed for DLP-based printing. Bioinks, including 15 wt% PEGDA (6 kDa)/25.5 mM LAP and 30 wt% PEGDA (6 kDa)/51 mM LAP, were developed for printing resolution tests. Sil-MA bioink composed of 15 wt% Sil-MA (EFL-Sil-MA-001, Yongqinquan Intelligent Equipment Co., Ltd., Suzhou, China) and 10 mM LAP was generated to verify the suitability of Cur-Na for other hydrogels.
Physico-chemical properties
Rheological properties of bioink were obtained using a MCR 102 parallel plate rheometer (Anton Par, Austria) equipped with an optics attachment where a 405 nm projector was adopted as the light source. 6 wt% PEGDA, 4 wt% GelMA and 17 mM LAP added with various concentrations of tartrazine and Cur-Na were used as the bioink formulation. The bioink specimens were loaded onto the plate, and the top fixture was adjusted to obtain a gap size of 200 μm. The sweep was performed every 3 min with a frequency of 1 Hz and 10% strain. The 405 nm light irradiation was initiated at 20 s and lasted for 30 s with an intensity of 13 mW cm−2. The time-series data of storage modulus G' and loss modulus G'' were then measured by RheoCompass software. A light exposure time of 30 s was selected because the resulting ∆G' log values (defined as the difference in the G' value of the crosslinked and uncrosslinked states) for PEG-GelMA/Cur-Na and PEG-GelMA/Tartrazine were ~78% and ~80% (see Fig. 2), respectively. Such values fall in the printable window (75–80% of ∆G' log values34), ensuring optimum crosslinking density.
Round-shaped samples (5 mm in diameter and 2 mm in depth) obtained by casting hydrogels into moulds were freeze-dried overnight to measure the dry weight (mdry t=0). The samples were then incubated overnight in adequate PBS at 37 °C to measure the wet weight (mswollen). Finally, the samples were further freezed-dried and weighed to obtain the dry weight (mdry). The swelling ratio can be calculated using the following equation7:
The sol fraction of the hydrogel was calculated as follows32:
The compressive stress–strain measurements were carried out using a tensile-compressive tester (Shimadzu-AGS-X with 20 N sensor). The round-shaped samples are 5 mm in diameter and 2 mm in height. The hydrogels were incubated in PBS at 37 °C for 4 h prior to the test. The compression rate was 1 mm min−1, and the compressive modulus was obtained by calculating the rate of stress–strain curves at the strain value of ~20%. All tests of physico-chemical properties were repeated three times.
Photopolymerisation kinetics
The polymerisation kinetics was characterised by measuring the conversion of C=C double bonds on real-time FT-IR spectroscopy (Nicolet iS10, Thermo Scientific, USA) equipped with a mercury cadmium telluride (MCT) detector. Samples (15 µL PEGDA or Curcumin-Na) were placed on an ATR crystal plate, and a 405 nm wavelength LED light was used for photopolymerising the materials. The light source was placed 3 cm above the sample, and the light intensity (13 mW cm−2) was calibrated and monitored using a Power Meter (PM16-401, Thorlabs, USA). The infrared absorbance spectra ranging from a wavenumber of 800–2000 cm−1 were recorded by Omnic software every 20 s for a total duration of 180 s (Supplementary Fig. 7), and the peak reflects the concentration of the double bonds. Each spectrum contains 16 scans with a resolution of 4 cm−1. The double bond conversion (α) was calculated as follows18:
where, At and A0 represent the absorption area at an irradiation time point and before light irradiation, respectively. At and A0 were obtained by computing the integration of the absorbance peak over 940–1000 cm−1 (see Supplementary Fig. 7).
3D printing
All structures to be printed were designed using the Computer-Aided Design (CAD) 2021 software and sliced using the BMF 3D slicer software. These structures were fabricated by a projection micro stereolithography (PμSL) printer (NanoArch S140, BMF Material Technology Inc, Shenzhen, China). The PμSL printer was equipped with a 405 nm projector with a resolution of 10 μm in the x–y plane. A light intensity of 13 mW cm−2 was used throughout the study, except for cases of light-intensity sensitivity study. After printing, the 3D-printed objects were washed with PBS for at least 3 min. Microscopy images of the fabricated parts were obtained using the Dino-Lite microscope with the DinoCapture 2.0 software.
Characterisation of the penetration depth
The relationship between the incident light intensity (E0) and curing depth (Cd) of bioink was calculated from the following equation1:
where Ec and Dp are the critical energy required to initiate polymerisation and the penetration depth related to the light absorbance of bioink materials, respectively. Cd and E0 were measured from experiments. Four circular specimens were cured with different exposure doses E0 varying between 156 and 312 mJ cm−2 (see Supplementary Fig. 6a), and the cured thickness Cd was measured using Dino-Lite digital microscope (AM4815ZT, Dino-Lite, China). Finally, Ec and Dp were obtained by linearly fitting the logarithmic plotting of measured Cd against E0 by using the equation above (see Supplementary Fig. 6b, c).
Characterisation of the printing resolution
A spoke-like pattern with sharp lines and an outer diameter of 5 mm (Fig. 4a) was designed and fabricated to evaluate the printing resolution of PEG-GelMA, PEG-GelMA/tartrazine and PEG-GelMA/Cur-Na. The printed structures were limited to a single layer thickness (100 μm) determined from the working curve in Supplementary Fig. 6. As the light energy to polymerise the three groups of bioink was different remarkably, the relative energy (Er) defined as the ratio between the actual light energy and the unit exposure dose was adopted for normalisation. The unit exposure dose was determined as the minimum light energy essential to solidify the spoke-like pattern properly. Five follow-up exposure doses Er = [1, 1.5, 2, 2.5, 3] were used to polymerise the spoke-like structures, and the printing resolution was assessed by comparing the unresolved fraction of the fabricated objects. For printing cell-laden structures, PC-12 cells were encapsulated in the bioink with a cell concentration of 1 × 107 mL−1. Fluorescence staining images were captured using Olympus inverted fluorescent microscope (IX-73, Olympus, Japan) with the Cellsens Standard software. The minimum printing resolution was tested via printing H letters designed with a linewidth of 1 pixel (10 μm). ImageJ was used to measure the printed structure’s dimensions (n = 3 per hydrogel).
Hollow-channel printability analysis
Cylindrical structures featuring open channels with diameters of 300, 500, and 700 μm (Fig. 5a) were used to evaluate the printing performance of the bioink by measuring the extent of blockage of the patterned channels. The structures with heights of 200, 600, 1000, 1500, 2000, or 3000 μm were fabricated using the DLP printer with a layer thickness of 100 μm. The unblocked fraction of the channel was measured using Olympus inverted fluorescent microscope (IX-73, Olympus, Japan). All printed parts were washed in PBS for 3 min before image acquisition.
Cells culture
PC-12 cells were initially induced with the nerve growth factor to obtain cells possessing neuronal characteristics. The PC-12 cells (from Procell Life Science&Technology Co., Ltd, China) were cultured in RPMI 1640 complete medium supplemented with 10% fetal bovine serum (Gibco Life Technologies, USA), 1% antibiotic solution (streptomycin, 100 μg mL−1, and penicillin, 100 units mL−1, Sigma-Aldrich Corp.). HepG2 cells (from Procell Life Science&Technology Co., Ltd, China) were cultured in high glucose Dulbecco’s modified Eagle’s medium (HG-DMEM) supplemented with 10% fetal bovine serum and 1% antibiotic solution. Culturing was performed in a humidified atmosphere incubator at 37 °C with 5% CO2 and 95% air.
Cell viability and proliferation
Cells were added to PEG-GelMA, PEG-GelMA/Tartrazine and PEG-GelMA/Cur-Na bioinks, forming a solution with a cell density of 5 × 105 mL−1. A series of concentrations, including 2 × 105, 5 × 105 and 1 × 106 mL−1, were tested. It was found that cells exhibited ideal viability and proliferation when a concentration of 5 × 105 mL−1 was selected. The printed cell-laden structures were cultured for 14 days in the above culture medium. Cytotoxicity was evaluated using a Live/dead assay kit (Beyotime Biotechnology, China) following the manufacturer’s instructions. Samples were visualised using a fluorescent microscope (IX-73, Olympus, Tokyo, Japan) after 30-min incubation at 37 °C and 5% CO2. The CCK-8 assays (Meilunbio, Dalian, China) were used to assess cell proliferation quantitatively (Thermo Scientific Multiskan FC), which was recorded by Skanlt RE 6.1.1 software.
Characterisation of cytocompatibility of 3D cell-laden scaffolds
The cell-laden scaffolds were fabricated using the PμSL 3D printing system under sterile conditions. The bioink was filtered using a 0.22 μm filter. The PC-12 or HepG2 cells were added to the PEG-GelMA/Cur-Na bioink, and the cell density of the solution was 1–2 × 107 mL−1. The temperature of the bioink tank and the building platform were maintained at ~37 °C. The cell-laden 3D scaffolds were cultured in the culture medium for 1, 5, 10 and 14 days to observe cell proliferation. The culture medium was changed daily. Samples were imaged using a fluorescent microscope (IX-73, Olympus, Tokyo, Japan) and a confocal microscope (Ti-E + A1 MP, Nikon, Tokyo, Japan) after 30-min incubation at 37 °C and 5% CO2. Confocal images were obtained using the Nikon Ti-E + A1 MP microscope with the NIS-Elements Viewer 4.50 software.
Albumin secretion
To evaluate the hepatic functions, the scaffolds culture supernatants were collected on days 1, 4, 7,10 and 14 and stored at −20 °C until use. The culture medium was changed daily, and at least three samples were used for each test. Albumin secretion was measured using the human albumin in vitro ELISA kit (SEKH-0081, Solarbio), and albumin concentration was calculated according to the experimental standard curve. Origin 2018 software was used for plotting.
Cell differentiation
To investigate the PC-12 cell differentiation, PC-12 cells were cultured on HG-DMEM supplemented with 10% fetal bovine serum, 1% antibiotic solution and 50 ng mL−1 nerve growth factor (NGF). The differentiation medium was refreshed daily. After 7 days of culturing, the differentiation of PC-12 cells was observed by immunostaining. The samples were washed twice with PBS. Then the samples were fixed in 4% formaldehyde solution for 15 min and washed thrice with PBS. The samples were soaked in 0.2% Triton-X 100 for 10 min and then blocked in PBST solution with 5% FBS for 1 h, followed by incubating overnight with the primary antibody of Anti-beta III tubulin (Tuj-1, 1:500, ab18207, Abcam) at 4 °C. The samples were then repeatedly washed with PBS to remove unbound antibodies and incubated for 1 h with goat anti-rabbit 546 (1:500, a11305, Invitrogen). Finally, the samples were counterstained with 4’,6-diaminyl-2-phenylindole (DAPI, 1:1, GTX30920, GeneTex) for 15 min. Samples were imaged using a fluorescence microscope (IX-73, Olympus).
Statistical analysis
The experimental results were present as mean values ± standard deviation (no less than three samples). The significant difference is determined by two-tailed t test, *P < 0.05, **P < 0.01, ***P < 0.001 and P > 0.05 (no significant difference). Significant differences between the means of parameters were calculated using the SPSS 17.0 software.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available in the paper and the Supplementary Information, and can be obtained upon request from the corresponding author fchen@hnu.edu.cn. Source data are provided together with this paper. Source data are provided with this paper.
References
Yu, C. et al. Photopolymerizable biomaterials and light-based 3D printing strategies for biomedical applications. Chem. Rev. 120, 10695–10743 (2020).
Mota, C., Camarero-Espinosa, S., Baker, M. B., Wieringa, P. & Moroni, L. Bioprinting: from tissue and organ development to in vitro models. Chem. Rev. 120, 10547–10607 (2020).
Murphy, S. V. & Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 32, 773–785 (2014).
Wang, Y., Kankala, R. K., Ou, C., Chen, A. & Yang, Z. Advances in hydrogel-based vascularised tissues for tissue repair and drug screening. Bioact. Mater. 9, 198–220 (2021).
Ozbolat, I. T., Peng, W. & Ozbolat, V. Application areas of 3D bioprinting. Drug Discov. Today 21, 1257–1271 (2016).
Khoon, S. L. et al. Fundamentals and applications of photo-cross-linking in bioprinting. Chem. Rev. 120, 10662–10694 (2020).
Kim, S. H. et al. Precisely printable and biocompatible silk fibroin bioink for digital light processing 3D printing. Nat. Commun. 9, 1620 (2018).
Ligon, S. C., Liska, R., Stampfl, J., Gurr, M. & Mulhaupt, R. Polymers for 3D printing and customized additive manufacturing. Chem. Rev. 117, 10212–10290 (2017).
Brett, E. K. et al. Volumetric additive manufacturing via tomographic reconstruction. Science 363, 1075–1079 (2019).
Grigoryan, B. et al. Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science 364, 458–464 (2019).
Xue, D. et al. Projection-based 3D printing of cell patterning scaffolds with multiscale channels. ACS Appl. Mater. Interfaces 10, 19428–19435 (2018).
Yu, F. & Choudhury, D. Microfluidic bioprinting for organ-on-a-chip models. Drug Discov. Today 24, 1248–1257 (2019).
Park, J., Wetzel, I., Dreau, D. & Cho, H. 3D miniaturization of human organs for drug discovery. Adv. Healthc. Mater. 7, 1700551 (2018).
Edmondson, R., Broglie, J. J., Adcock, A. F. & Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol 12, 207–218 (2014).
Antoni, D., Burckel, H., Josset, E. & Noel, G. Three-dimensional cell culture: a breakthrough in vivo. Int. J. Mol. Sci. 16, 5517–5527 (2015).
Fang, Y., Sun, W., Zhang, T. & **ong, Z. Recent advances on bioengineering approaches for fabrication of functional engineered cardiac pumps: a review. Biomaterials 280, 121298 (2022).
Huh, J. et al. Combinations of photoinitiator and UV absorber for cell-based digital light processing (DLP) bioprinting. Biofabrication 13, 034103 (2021).
Zhao, X. et al. Efficient 3D printing via photooxidation of ketocoumarin based photopolymerisation. Nat. Commun. 12, 2873 (2021).
You, S., Wang, P., Wang, P., Hwang, H. H. & Chen, S. High-fidelity 3D printing using flashing photopolymerization. Addit. Manuf. 30, 100834 (2019).
Pepelanova, I., Kruppa, K., Scheper, T. & Lavrentieva, A. Gelatin-methacryloyl (GelMA) hydrogels with defined degree of functionalization as a versatile toolkit for 3D cell culture and extrusion bioprinting. Bioengineering 5, 55 (2018).
Zhu, W. et al. Rapid continuous 3D printing of customisable peripheral nerve guidance conduits. Mater. Today 21, 951–959 (2018).
Li, H., Tan, Y. J., Kiran, R., Tor, S. B. & Zhou, K. Submerged and non-submerged 3D bioprinting approaches for the fabrication of complex structures with the hydrogel pair GelMA and alginate/methylcellulose. Addit. Manuf. 37, 101640 (2021).
Koffler, J. et al. Biomimetic 3D-printed scaffolds for spinal cord injury repair. Nat. Med. 25, 263–269 (2019).
Melchels, F. P., Feijen, J. & Grijpma, D. W. A review on stereolithography and its applications in biomedical engineering. Biomaterials 31, 6121–6130 (2010).
Guan, J. et al. Compensating the cell-induced light scattering effect in light-based bioprinting using deep learning. Biofabrication 14, 015011 (2021).
Ferrage, L., Bertrand, G., Lenormand, P., Grossin, D. & Ben-Nissan, B. A review of the additive manufacturing (3DP) of bioceramics: alumina, zirconia (PSZ) and hydroxyapatite. J. Aust. Ceram. Soc. 53, 11–20 (2016).
Zakeri, S., Vippola, M. & Levänen, E. A comprehensive review of the photopolymerisation of ceramic resins used in stereolithography. Addit. Manuf. 35, 101177 (2020).
Stevens, L. J., Burgess, J. R., Stochelski, M. A. & Kuczek, T. Amounts of artificial food dyes and added sugars in foods and sweets commonly consumed by children. Clin. Pediatr. 54, 309–321 (2015).
Lim, K. S. et al. Bio-resin for high resolution lithography-based biofabrication of complex cell-laden constructs. Biofabrication 10, 034101 (2018).
Simon, U. & Dimartino, S. Direct 3D printing of monolithic ion exchange adsorbers. J. Chromatogr. A 1587, 119–128 (2019).
Ouyang, X. et al. Optical micro-printing of cellular-scale microscaffold arrays for 3D cell culture. Sci. Rep. 7, 8880 (2017).
Levato, R. et al. High-resolution lithographic biofabrication of hydrogels with complex microchannels from low-temperature-soluble gelatin bioresins. Mater. Today Bio 12, 100162 (2021).
Raman, R. et al. High-resolution projection microstereolithography for patterning of neovasculature. Adv. Healthc. Mater. 5, 610–619 (2016).
Yu, K. et al. Printability during projection-based 3D bioprinting. Bioact. Mater. 11, 254–267 (2021).
Yin, J., Yan, M., Wang, Y., Fu, J. & Suo, H. 3D bioprinting of low-concentration cell-laden gelatin methacrylate (GelMA) bioinks with a two-step cross-linking strategy. ACS Appl. Mater. Interfaces 10, 6849–6857 (2018).
You, S. et al. Mitigating scattering effects in light-based three-dimensional printing using machine learning. J. Manuf. Sci. Eng 142, 081002 (2020).
Wang, Z. et al. A novel, well-resolved direct laser bioprinting system for rapid cell encapsulation and microwell fabrication. Adv. Healthc. Mater. 7, e1701249 (2018).
**e, M. et al. Electro-assisted bioprinting of low-concentration GelMA microdroplets. Small 15, e1804216 (2019).
Cha, C. et al. Structural reinforcement of cell-laden hydrogels with microfabricated three dimensional scaffolds. Biomater. Sci. 2, 703–709 (2014).
Chen, Y. C. et al. Functional human vascular network generated in photocrosslinkable gelatin methacrylate hydrogels. Adv. Funct. Mater. 22, 2027–2039 (2012).
Choi, J. R., Yong, K. W., Choi, J. Y. & Cowie, A. C. Recent advances in photo-crosslinkable hydrogels for biomedical applications. BioTechniques 66, 40–53 (2019).
Khoshakhlagh, P., Bowser, D. A., Brown, J. Q. & Moore, M. J. Comparison of visible and UVA phototoxicity in neural culture systems micropatterned with digital projection photolithography. J. Biomed. Mater. Res. A 107, 134–144 (2019).
Ravve, A. Principles of Polymer Chemistry (Springer, 2012).
Li, Y. et al. High-fidelity and high-efficiency additive manufacturing using tunable pre-curing digital light processing. Addit. Manuf. 30, 100889 (2019).
Liu, X. et al. 3D bioprinted neural tissue constructs for spinal cord injury repair. Biomaterials 272, 120771 (2021).
Li, P., Xu, Y., Cao, Y. & Wu, T. 3D digital anatomic angioarchitecture of the rat spinal cord: a synchrotron radiation micro-CT study. Front. Neuroanat. 14, 41 (2020).
Fang, Y. et al. Optimizing bifurcated channels within an anisotropic scaffold for engineering vascularized oriented tissues. Adv. Healthc Mater. 9, e2000782 (2020).
Zhu, W. et al. Direct 3D bioprinting of prevascularized tissue constructs with complex microarchitecture. Biomaterials 124, 106–115 (2017).
Han, X. et al. Optimized vascular network by stereolithography for tissue engineered skin. Int. J. Bioprint. 4, 134 https://doi.org/10.18063/IJB.v4i2.134 (2018).
Han, X., Bibb, R. & Harris, R. Engineering design of artificial vascular junctions for 3D printing. Biofabrication 8, 025018 (2016).
Feng, J., Fu, J., Yao, X. & He, Y. Triply periodic minimal surface (TPMS) porous structures: from multiscale design, precise additive manufacturing to multidisciplinary applications. Int. J. Extrem. Manuf. 4, 022001 (2022).
Zhang, Q. et al. High‐strength hydroxyapatite scaffolds with minimal surface macrostructures for load‐bearing bone regeneration. Adv. Funct. Mater. 32, 2204182 (2022).
Jung, M., Ghamrawi, S., Du, E. Y., Gooding, J. J. & Kavallaris, M. Advances in 3D bioprinting for cancer biology and precision medicine: from matrix design to application. Adv. Healthc. Mater. 11, e2200690 (2022).
Germain, L., Fuentes, C. A., van Vuure, A. W., des Rieux, A. & Dupont-Gillain, C. 3D-printed biodegradable gyroid scaffolds for tissue engineering applications. Mater. Design 151, 113–122 (2018).
Miller, J. S. et al. Bioactive hydrogels made from step-growth derived PEG-peptide macromers. Biomaterials 31, 3736–3743 (2010).
Shirahama, H., Lee, B. H., Tan, L. P. & Cho, N.-J. Precise tuning of facile One-pot gelatin methacryloyl (GelMA) synthesis. Sci. Rep. 6, 31036 (2016).
Fairbanks, B. D., Schwartz, M. P., Bowman, C. N. & Anseth, K. S. Photoinitiated polymerisation of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerisation rate and cytocompatibility. Biomaterials 30, 6702–6707 (2009).
Acknowledgements
X.H. acknowledges the financial support received from the National Natural Science Foundation of China (52075158), the National Natural Science Foundation of Hunan Province (2021JJ30109) and the Innovative Platform and Talents Program of Hunan Province (2019RS1019). X.H. and F.C. acknowledge the financial support received from the Key R&D Program of Hunan Province (2020SK2093). N.H. is grateful for the financial support from the Postgraduate Scientific Research Innovation Project of Hunan Province (CX20220407). The authors also thank Mr. Haotian Zhu (an undergraduate student at Hunan University) for the CAD design of the spinal cord scaffold.
Author information
Authors and Affiliations
Contributions
N.H.: methodology, formal analysis, data curation, validation, and original draft writing; X.W. and L.S.: biological resources, formal analysis, and manuscript review; Y.H. and H.L.: material resources and experimental analysis; J.L. and X.Z.: visualization, data curation, and methodology; L.M.: material synthesis and formal analysis; Y.M. and W.Z.: software, investigation, and manuscript review; F.C. and X.H.: conceptualisation, investigation, project administration, review and editing, supervision and funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Chan Hum Park and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
He, N., Wang, X., Shi, L. et al. Photoinhibiting via simultaneous photoabsorption and free-radical reaction for high-fidelity light-based bioprinting. Nat Commun 14, 3063 (2023). https://doi.org/10.1038/s41467-023-38838-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-023-38838-2
- Springer Nature Limited
This article is cited by
-
3D Bioprinting of Cultured Meat: A Promising Avenue of Meat Production
Food and Bioprocess Technology (2024)