Abstract
The nuclear envelope (NE) is often challenged by various stresses (known as “NE stress”), leading to its dysfunction. Accumulating evidence has proven the pathological relevance of NE stress in numerous diseases ranging from cancer to neurodegenerative diseases. Although several proteins involved in the reassembly of the NE after mitosis have been identified as the NE repair factors, the regulatory mechanisms modulating the efficiency of NE repair remain unclear. Here, we showed that response to NE stress varied among different types of cancer cell lines. U251MG derived from glioblastoma exhibited severe nuclear deformation and massive DNA damage at the deformed nuclear region upon mechanical NE stress. In contrast, another cell line derived from glioblastoma, U87MG, only presented mild nuclear deformation without DNA damage. Time-lapse imaging demonstrated that repairing of ruptured NE often failed in U251MG, but not in U87MG. These differences were unlikely to have been due to weakened NE in U251MG because the expression levels of lamin A/C, determinants of the physical property of the NE, were comparable and loss of compartmentalization across the NE was observed just after laser ablation of the NE in both cell lines. U251MG proliferated more rapidly than U87MG concomitant with reduced expression of p21, a major inhibitor of cyclin-dependent kinases, suggesting a correlation between NE stress response and cell cycle progression. Indeed, visualization of cell cycle stages using fluorescent ubiquitination-based cell cycle indicator reporters revealed greater resistance of U251MG to NE stress at G1 phase than at S and G2 phases. Furthermore, attenuation of cell cycle progression by inducing p21 in U251MG counteracted the nuclear deformation and DNA damage upon NE stress. These findings imply that dysregulation of cell cycle progression in cancer cells causes loss of the NE integrity and its consequences such as DNA damage and cell death upon mechanical NE stress.
Similar content being viewed by others
Introduction
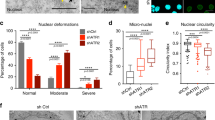
The nuclear envelope (NE) encloses genomic DNA to protect it from a variety of insults and plays pivotal roles in regulating genomic function [1, 2]. Previous studies demonstrated that the NE is damaged upon various types of stresses, leading to its dysfunction [3,4,5,6,7]. Such stresses are referred to as “NE stress” [8,9,10,11]. Mechanical NE stress frequently results in the rupture of the NE [3, 4]. Cell migration through confined environments is one of the major causes of NE rupture [3, 4]. This can include both physiological and pathological events such as the radial migration of neural progenitors during brain development and the invasion of tumor cells into surrounding tissues, respectively [3, 4, 12]. Accumulating evidence has revealed that NE rupture is involved in a broad spectrum of diseases ranging from cancer to neurodegenerative diseases. Regarding the pathology of cancer, NE rupture has been proposed to trigger genome instability and increased cellular invasion through DNA damage [13, 14]. Mechanistically, studies have suggested that the mislocalization of cytoplasmic DNase and/or nuclear DNA repair factors is responsible for such DNA damage [14, 12]. These findings imply that the consequences of NE stress might be context dependent. Thus, we investigated whether mechanical NE stress causes apoptosis in our model. To achieve this, U251MG, U87MG, and NP5 were plated on a Transwell, and apoptosis was detected by immunostaining using anti-cleaved caspase-3 antibodies. Substantial cleaved caspase-3 signals were observed at the bottom side of the Transwell for U251MG and NP5, whereas for U87MG they were hardly detected (Fig. 7A, B). These findings suggest that mechanical NE stress results in the apoptotic cell death of U251MG and NP5, presumably due to inefficient repair of ruptured NE. To test whether p21 plays pivotal roles in the suppression of NE stress-mediated apoptosis, U87MG cells were subjected to a Transwell assay with and without depletion of p21. Depletion of p21 resulted in a significant increase of the presence of the clusters of cleaved-caspase-3 (Fig. 7C, D). Thus, it is suggested that p21 is indispensable for the suppression of apoptosis induced by NE stress in U87MG cells.
A Immunofluorescence staining analysis of cleaved caspase-3 (green) of U251MG, U87MG, and NP5 at the bottom side of a Transwell. Overlay indicates merged images with DNA (blue). Scale bars: 10 μm. B Quantification of cleaved caspase-3 clusters relative to total cell number in the cells indicated in (A). Bars and error bars represent the mean values and standard deviations from three independent experiments. Statistical significance of the differences was determined using Fisher’s exact test. ***p < 0.005. Total cell numbers for each cell line from three independent experiments: n = 1045, U251MG; n = 701, U87MG; n = 357, NP5. C Immunofluorescence staining analysis of cleaved caspase-3 (green) of U87MG cells with (sip21) or without (siCon) RNAi of p21 at the bottom side of a Transwell. Overlays represent merged images with DNA (blue). Scale bars: 5 μm. D Quantification of cleaved caspase-3 clusters relative to total cell number in the cells indicated in (C). Bars and error bars represent the mean values and standard deviations from three independent experiments. Statistical significance of the differences was determined using Fisher’s exact test. *p < 0.05. Total cell numbers for each condition from three independent experiments: n = 540, siCon; n = 1055, sip21.
Discussion
While some of the molecular mechanisms of NE stress response have been clarified, how such processes are affected by the types of cells and/or the cellular environment has not been elucidated. In the present study, we demonstrated that the response to mechanical NE stress varies among cancer cell lines. Furthermore, our data revealed the impact of the cell cycle on sensitivity to mechanical NE stress in GBM. Mechanistically one of the major CDK inhibitors, p21, is indispensable for the resistance to mechanical NE stress and suppresses the consequences of NE stress.
In higher eukaryotes, the NE is completely disassembled at the early onset of mitosis to allow access to the mitotic spindle to the chromosomes [22]. Thus, disassembly and subsequent reassembly of the NE are essential for the progression of the cell cycle. Prior to the disassembly of the NE, many NE components, including BAF and LEM domain proteins, are dissociated from the NE [22]. The change of their localization is mainly regulated by cell-cycle-dependent phosphorylation through several kinases including CDK1 [22, 24, 25, 32]. After the segregation of chromosomes, NE components are recruited to the surface of the chromosomes to establish functional NE. In contrast to the disassembly of the NE, reassembly requires the dephosphorylation of many NE proteins, which is mediated by the inactivation of mitotic kinases [22]. Currently, BAF is known as the earliest accumulating factor to the chromosome among the proteins that are dissociated from the NE prior to NE disassembly [22, 23, 33]. BAF targets LEM domain proteins, leading to further recruitment of ESCRT III complex to coordinate reseal the NE and remove the spindle from chromosomes [22, 34]. Some studies have proposed that the repair of ruptured NE involves a process similar to NE reassembly, featuring sequential targeting of BAF, LEM domain proteins, and ESCRT III to the ruptured site [16, 17, 23]. Thus, it is highly possible that the regulatory mechanisms are also shared between the repair and reassembly of the NE. In this scenario, phosphorylation of the NE repair factors related to the disassembly of the NE will perturb the efficiency of NE repair due to the loss of their capacity to accumulate at the ruptured NE. This can explain why G2-phase of cells are more sensitive to NE stress even if the expression of p21 is absent in the entire population of U251MG cells. In this regard, p21 can restore effective NE repair by inhibiting the phosphorylation of NE repair factors. Furthermore, because p21 is depleted in the S phase in U87MG cells, sensitivity to mechanical NE stress is possibly higher at this stage of cell cycle in this cell line as well as in other cancer cells with the same pattern of p21 expression. Dysregulation of the cell cycle is frequently observed in many types of cancer, which potentially impairs the efficient repair of ruptured NE [35]. Our data further support this idea and reveal the link between the cell cycle and NE stress response.
It is possible that cell-cycle-dependent perturbation of NE repair occurs not only in GBM but also in other types of cancers, although we have not observed severe consequences of mechanical NE stress in those cancer cell lines (Fig. 1). Additional investigations will clarify whether the unknown cell-cycle-independent NE repair pathway plays pivotal roles to maintain NE integrity upon mechanical NE stress in those cells. Moreover, further studies are also required to elucidate the significance of apoptosis caused by NE stress in NE stress sensitive cells. Regarding the malignancy of GBM, it is possible that G1-phase cells are responsible for the invasion of the GBM in vivo due to their resistance to mechanical NE stress.
In summary, this study revealed that (1) the efficiency of repair of ruptured NE is regulated in a cell-cycle-dependent manner and (2) impaired NE repair caused by dysregulation of cell cycle triggers severe consequences of NE stress in GBM. These novel insights indicate that further detailed investigation of the molecular mechanisms behind the regulation of NE repair should make significant contributions to the development of an innovative therapeutic strategy of GBM.
Materials and methods
Cell culture, plasmids, and stable cell lines
U251MG, HT1080, DU145, A549, and U87MG cells were maintained in Eagle’s Minimum Essential Medium (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS), 1 mM sodium pyruvate, 1X non-essential amino acids, and penicillin/streptomycin at 37 °C in a 5% CO2, 95% humidified air atmosphere. MDA-MB468 cells were maintained in Leibovitz’s L-15 Medium (FUJIFILM Wako Pure Chemical Corporation) supplemented with 10% FBS and penicillin/streptomycin at 37 °C in 100% air atmosphere. NP5 cells were maintained in Dulbecco’s Modified Eagle Medium (FUJIFILM Wako Pure Chemical Corporation) supplemented with 10% FBS and penicillin/ streptomycin at 37 °C in 5% CO2, 95% humidified air atmosphere. Stable cell lines were established using either transposon or lentivirus. Plasmids for Slee**beauty based transposon vectors were obtained from addgene. Donor plasmids (pSBbi-Bla: #60526; pSBTet-Pur: # 60507) were transfected together with the expression plasmid for a mutant form of transposase, SB100X (pCMV(CAT)T7-SB100: #34879) using Everyday Transfection (EZ Biosystems, College Park, MD, USA) according to manufacturer’s protocol [36, 37]. A NLS derived from SV40 large T antigen was fused with N-terminus of tdTomato by PCR and resulting fragment was cloned into pSBbi-Bla vector (amplified with PCR) using In-Fusion system (TaKaRa Bio, Kusatshu, Shiga, Japan). cDNA of human p21 was obtained by reverse transcription using ReverTra Ace (TOYOBO, Osaka, Japan) and RNA purified from U87MG. PCR was performed to clone p21 into pmVenus-C1 using BsrGI/EcoRI sites [9]. Then, resulting mVenus-p21 was amplified with PCR and cloned into pSBTet-Pur (digested with SfiI) using In-Fusion system. The stable cell lines expressing mScarlet-BAF were generated by transposon-based vectors indicated above. cDNA of BAF is amplified by PCR using EGFP-BAF plasmid (addgene number: 52962) and cloned into pSBbi-Bla together with mScarlet-I (a gift from Dorus Gadella, addgene number: 98831) [38]. The primers used for construction are listed in Supplementary Table 1. Integrated cells were selected using blasticydin (8 μg/ml) or puromycin (2 μg/ml). The plasmid for FUCCI was obtained from RIKEN BRC (#RDB15454) [30]. To introduce FUCCI reporter set, pCSII lentivirus vector carrying mCherry-hCdt1(1/100)Cy(-) and mTurquoise-hGem(1/110) was transfected into HEK293T cells together with LP1, LP2, and VSV-G plasmids. Resulting lentivirus particles were concentrated using Lenti-X Concentrator (TaKaRa Bio) according to manufacturer’s protocol. Polybrene (8 μg/ml) was supplemented to the medium when cells were infected with lentiviruses.
Constricted migration assay using Transwell
Constricted migration assay was performed using Transwell with a 3 μm pore size (Corning Incorporated, Corning, NY, USA) as described previously with minor modifications [9]. 1 × 105 cells were plated on a Transwell and cultured for 14–16 h, followed by immunofluorescence staining. For immunofluorescence staining of cleaved-caspase-3, cell were cultured for 40 h.
EdU labeling and click reaction
To identify S-phase cells, EdU (Jena Bioscience, Jena, Thuringia, Germany) was added into culture medium at 25 μM and incubated for 30 min. Incorporated EdU was visualized by a fluorescent dye through a Cu(I)-catalyzed [3 + 2] cycloaddition (click) reaction, by which terminal alkyne group of EdU is covalently attached with azide group of a fluorescent dye, as described previously with minor modifications [39, 40]. Briefly, cells were fixed with 4% paraformaldehyde (PFA) for 15 min and permeabilized with 0.5% Triton X-100 for 3 min. After three times PBS washes, cells were incubated with the reaction mixture (2 μM picolyl azide Alexa-555 (Jena Bioscience), 20 mM Sodium ascorbate (FUJIFILM Wako Pure Chemical Corporation), 2 mM 2-(4-((bis((1-(tert-butyl)-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl)acetic acid (Jena Bioscience), 1 mM CuSO4 (FUJIFILM Wako Pure Chemical Corporation) diluted in PBS) for 30 min at 37 °C. After three times PBS wash, cells were subjected to the immunostaining if necessary.
Immunofluorescence staining and nuclear circularity
The cells were grown on coverslips (Matsunami Glass, Osaka, Japan) or Transwell and fixed in 4% PFA for 15 min. After three times wash with PBS, the cells were permeabilized in 0.5% Triton‐X 100 for 3 min followed by blocking with 10% normal goat serum for 60 min. The following antibodies were used as the primary antibodies: anti‐lamin B1 (1:1000; ab16048, Abcam, Cambridge, UK), anti-γH2A.X (1:1000; 05-636, Merk Millipore, Burlington, MA, USA), anti-cyclin B1 (1:200; SC-245, Santa Cruz Biotechnology, Dallas, TX, USA) anti-p21 (1:500; #2947, Cell Signaling Technology, Danvers, MA, USA), anti-cleaved caspase-3 (1:500; #9661, Cell Signaling Technology). Incubation with primary antibodies was performed at 4 °C overnight or 2 h at R.T. For secondary antibodies, goat anti-mouse or anti-rabbit IgG F(ab′)2 fragments conjugated with Alexa 488 or 568 were used (Thermo Fisher Scientific, Waltham, MA, USA). Confocal fluorescent images were acquired using FV1000D (Evident, Tokyo, Japan), FV3000 (Evident, Tokyo, Japan), or Stellaris5 (Leica Microsystems, Wetzlar, Germany). All images were processed through imageJ (National Institutes of Health, Rockville, MD, USA) [41]. The prominent signal of γH2A.X is defined manually with same setting of contrast among a series of the experiments. Obtained images of DNA staining using DAPI were processed to measure nuclear circularity as bellow using imageJ. All the images were subjected to “Gaussian Bluer (sigma 1)” and then Z-stacked with maximum intensity. The binary images were generated using “Threshold (Li, auto)” and subjected to “Fill holes” filter. Then, the nuclear circularity was measured using “Find particles”. Overlapped nuclei were manually excluded.
Laser irradiation
U251MG and U87MG cells expressing NLS-tdTomato or mScarlet-BAF were plated on the glass bottom dish (Matsunami glass, Osaka, Japan) coated with rat collagen I (Thermo Scientific). Laser irradiation and time lapse imaging was performed using FV1000D. A circular spot with 0.6 μm diameter at the periphery of the nucleus was irradiated with a 405 nm laser at 100% power for 6 s using a SIM light pathway. DNA was stained with SPY650 DNA probe (SPIROCHROME, Thurgau, Switzerland) and visualized with a 635 nm laser. Time lapse imaging was performed with 3 min intervals for 57 min. Obtained images of NLS-tdTomato were processed to measure the ratio of NLS-tdTomato signal in cytoplasm to the nucleus as bellow using imageJ. All the images were subjected to “Subtract Background (Rolling ball radius: 50 pixels, Sliding paraboloid, Disable smoothing), “Gaussian Bluer (sigma 1)”, and then Z-stacked with maximum intensity. Some regions of the nucleus and cytoplasm were manually selected and the mean signal intensity were measured. The signal intensity of cytoplasmic NLS-tdTomato was divided by that of the nucleus, and its highest values during the time course was set as 1.0 for each cell. For the immunostaining of γH2A.X after laser irradiation, the cells were cultured additional 16 h and subjected to immunostaining. All the regions that possibly include irradiated cells were visualized with tile scan using Stellaris5.
Protein preparation and western blotting
Proteins were extracted from the indicated cells in RIPA buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% SDS, 0.5% Sodium deoxycholate, 1% Triton X-100) supplemented with Protease inhibitor cocktail Set V/Phosphatase inhibitor cocktail Set (FUJIFILM Wako Pure Chemical Corporation). The lysates were incubated for 30 min on ice. After centrifugation at 15,000xg for 5 min, the protein concentrations of the supernatants were determined using a bicinchoninic acid assay kit (Thermo Fisher Scientific). Equal amounts of proteins were used for SDS-PAGE. For immunoblotting, the following antibodies were used: anti-cyclin B1 (1:200; SC-245, Santa Cruz Biotechnology, Dallas, TX USA), anti-cyclin E (1:200; SC-247, Santa Cruz), anti-p21 (1:2000; #2947, Cell Signaling Technology), anti‐lamin A/C (1:2000; MABT1341, Merk Millipore), anti-MAN1 (1:10000; TA500794, OriGene, Rockville, MD USA) anti‐β‐actin (1:5000; MAB1501, Merk Millipore).
Flow cytometry
The cells were trypsinized, washed with PBS once, and fixed with 70% of EtOH for 30 min at 4 °C. After PBS wash, the cells were stained with PI solution (0.15 % Triton X-100, 2μg/ml PI, 50μg/ml RNaseA) at R.T. for 30 min. Cell cycle profile was obtained using FlowJo.
RNA interference
For knockdown of p21, small interfering (si)RNA targeted to human p21 (AM51331) and negative control (AM4611) were purchased from Thermo Fisher Scientific. siRNA were transfected using Lipofectamine RNAiMax (Thermo Fisher Scientific) as described in the manufacture’s manual. The depletion of p21 was confirmed by western blot 48 h after transfection. Transfection was performed on the top side of the Transwell without the medium at the bottom side. After 12 h Transfection, the medium was added to the bottom side, then the cells were cultured additional 36 h before immunostaining.
Statistical analysis
Microsoft Excel and R (version 4.0.2) were used via RStudio (version 1.3.1073) for statistical analysis [42, 43].
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol. 2010:2:a000539.
Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol. 2010;11:317–28.
Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, et al. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–8.
Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–62.
De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, et al. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum Mol Genet. 2011;20:4175–86.
de Noronha CM, Sherman MP, Lin HW, Cavrois MV, Moir RD, Goldman RD, et al. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science. 2001;294:1105–8.
Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 2012;3:88–100.
Di Micco A, Frera G, Lugrin J, Jamilloux Y, Hsu E-T, Tardivel A, et al. AIM2 inflammasome is activated by pharmacological disruption of nuclear envelope integrity. Proc Natl Acad Sci USA. 2016;113:E4671–4680.
Kamikawa Y, Saito A, Matsuhisa K, Kaneko M, Asada R, Horikoshi Y, et al. OASIS/CREB3L1 is a factor that responds to nuclear envelope stress. Cell Death Discov. 2021;7:152.
Robijns J, Houthaeve G, Braeckmans K, De Vos WH. Loss of nuclear envelope integrity in aging and disease. Int Rev Cell Mol Biol. 2018;336:205–22.
Kamikawa Y, Imaizumi K. Advances in understanding the mechanisms of repairing damaged nuclear envelope. J Biochem. 2022;171:609–17.
Chen NY, Yang Y, Weston TA, Belling JN, Heizer P, Tu Y, et al. An absence of lamin B1 in migrating neurons causes nuclear membrane ruptures and cell death. Proc Natl Acad Sci USA. 2019;116:25870–9.
Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and kataegis induced by telomere crisis. Cell. 2015;163:1641–54.
de Nader GPF, Agüera-Gonzalez S, Routet F, Gratia M, Maurin M, Cancila V, et al. Compromised nuclear envelope integrity drives TREX1-dependent DNA damage and tumor cell invasion. Cell. 2021;184:5230–46.e22.
Irianto J, **a Y, Pfeifer CR, Athirasala A, Ji J, Alvey C, et al. DNA damage follows repair factor depletion and portends genome variation in cancer cells after pore migration. Curr Biol. 2017;27:210–23.
Halfmann CT, Sears RM, Katiyar A, Busselman BW, Aman LK, Zhang Q, et al. Repair of nuclear ruptures requires barrier-to-autointegration factor. J Cell Biol. 2019;218:2136–49.
Young AM, Gunn AL, Hatch EM. BAF facilitates interphase nuclear membrane repair through recruitment of nuclear transmembrane proteins. Mol Biol Cell. 2020;31:1551–60.
Kono Y, Adam SA, Sato Y, Reddy KL, Zheng Y, Medalia O, et al. Nucleoplasmic lamin C rapidly accumulates at sites of nuclear envelope rupture with BAF and cGAS. J Cell Biol. 2022;221:e202201024.
Olmos Y, Perdrix-Rosell A, Carlton JG. Membrane ninding by CHMP7 coordinates ESCRT-III-dependent nuclear envelope reformation. Curr. Biol. 2016;26:2635–41.
Vietri M, Schultz SW, Bellanger A, Jones CM, Petersen LI, Raiborg C, et al. Unrestrained ESCRT-III drives micronuclear catastrophe and chromosome fragmentation. Nat Cell Biol. 2020;22:856–67.
Coyne AN, Rothstein JD. The ESCRT-III protein VPS4, but not CHMP4B or CHMP2B, is pathologically increased in familial and sporadic ALS neuronal nuclei. Acta Neuropathol Commun. 2021;9:127.
Ungricht R, Kutay U. Mechanisms and functions of nuclear envelope remodeling. Nat Rev Mol Cell Biol. 2017;18:229–45.
Maciejowski J, Hatch EM. Nuclear membrane rupture and its consequences. Annu Rev Cell Dev Biol. 2020;36:85–114.
Nichols RJ, Wiebe MS, Traktman P. The vaccinia-related kinases phosphorylate the N’ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol Biol Cell. 2006;17:2451–64.
Hirano Y, Segawa M, Ouchi FS, Yamakawa Y, Furukawa K, Takeyasu K, et al. Dissociation of emerin from barrier-to-autointegration factor is regulated through mitotic phosphorylation of emerin in a xenopus egg cell-free system. J Biol Chem. 2005;280:39925–33.
Shimi T, Pfleghaar K, Kojima S, Pack C-G, Solovei I, Goldman AE, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–21.
Hatch EM, Hetzer MW. Nuclear envelope rupture is induced by actin-based nucleus confinement. J Cell Biol. 2016;215:27–36.
Harada T, Swift J, Irianto J, Shin J-W, Spinler KR, Athirasala A, et al. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol. 2014;204:669–82.
Harper JW, Elledge SJ, Keyomarsi K, Dynlacht B, Tsai LH, Zhang P, et al. Inhibition of cyclin-dependent kinases by p21. MBoC. 1995;6:387–400.
Sakaue-Sawano A, Yo M, Komatsu N, Hiratsuka T, Kogure T, Hoshida T, et al. Genetically encoded tools for optical dissection of the mammalian cell cycle. Mol Cell. 2017;68:626–40.e5.
Depmap: Cancer Dependency Map: https://depmap.org/portal/.
Gatta AT, Olmos Y, Stoten CL, Chen Q, Rosenthal PB, Carlton JG. CDK1 controls CHMP7-dependent nuclear envelope reformation. Elife. 2021;10:e59999.
Haraguchi T, Kou** T, Segura-Totten M, Lee KK, Matsuoka Y, Yoneda Y, et al. BAF is required for emerin assembly into the reforming nuclear envelope. J Cell Sci. 2001;114:4575–85.
von Appen A, LaJoie D, Johnson IE, Trnka MJ, Pick SM, Burlingame AL, et al. LEM2 phase separation promotes ESCRT-mediated nuclear envelope reformation. Nature. 2020;582:115–8.
Suski JM, Braun M, Strmiska V, Sicinski P. Targeting cell-cycle machinery in cancer. Cancer Cell. 2021;39:759–78.
Kowarz E, Löscher D, Marschalek R. Optimized Slee** Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J. 2015;10:647–53.
Mátés L, Chuah MKL, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, et al. Molecular evolution of a novel hyperactive Slee** Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–61.
Chertkova AO, Mastop M, Postma M, van Bommel N, van der Niet S, Batenburg KL, et al. Robust and bright genetically encoded fluorescent markers for highlighting structures and compartments in mammalian cells. 160374. https://doi.org/10.1101/160374 (2020).
Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–20.
Lee SR, Pollard DA, Galati DF, Kelly ML, Miller B, Mong C, et al. Disruption of a ∼23–24 nucleotide small RNA pathway elevates DNA damage responses in Tetrahymena thermophila. MboC. 2021;32:1335–46.
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82.
R Core Team. R: A Language and Environment for Statistical Computing 2020. R Foundation for Statistical Computing, Vienna, Austria. Available from https://www.R-project.org/.
Rstudio Team. Rstudio: Integrated Development for R. 2020. Rstudio, PBC, Boston, MA. Available from http://www.rstudio.com/.
Acknowledgements
The authors thank Eric Kowarz, Zsuzsanna Izsvak, Dorus Gadella, Daniel Gerlich, and Atsushi Miyawaki for providing their plasmids. This work was partly supported by grants from the Japan Society for the Promotion of Science KAKENHI (22K06129, 20K07573), GUSINKAI FOUNDATION, the Program of the Network-type Joint Usage/Research Center for Radiation Disaster Medical Science, Core Research for Organelle Diseases in Hiroshima University (the MEXT program for promoting the enhancement of research universities, Japan), Takeda Science Foundation, and Joint Usage and Joint Research Programs, the Institute of Advanced Medical Sciences, Tokushima University. The editing the English text of a draft of this manuscript has been done by Edanz. Some of the equipment used in this study was provided by the Natural Science Center for Basic Research and Development (NBARD-00007), Hiroshima University.
Author information
Authors and Affiliations
Contributions
YK and AS developed the tools for experiments. YK and ZW performed experiments. NN and TI supported the analysis of public available data using DepMap. KI supervised the project. YK and KI designed the experiments, analyzed data, and wrote the manuscript. AS provided input into conceptualizing the project and writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kamikawa, Y., Wu, Z., Nakazawa, N. et al. Impact of cell cycle on repair of ruptured nuclear envelope and sensitivity to nuclear envelope stress in glioblastoma. Cell Death Discov. 9, 233 (2023). https://doi.org/10.1038/s41420-023-01534-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-023-01534-7
- Springer Nature Limited