Abstract
Accumulating evidence has shown that circular RNA (circRNA) dysregulation is involved in various types of cancer, including osteosarcoma (OS). Nevertheless, the role and mechanism of circRNAs in OS progression and chemoresistance remain elusive. We found that a novel doxorubicin-induced circular RNA, hsa_circ_0004674, screened by whole total transcriptome RNA sequencing in our previous study, was upregulated in OS chemoresistant cell lines and tissues and also connected with patients’ poor prognosis. Circ_0004674 knockdown remarkably suppressed OS cell chemoresistance, proliferation, migration, invasion, OS tumor growth, and enhanced cell cycle arrest and apoptosis in vitro and in vivo through control the expression of the antiapoptotic protein MCL1, a member of the Bcl-2 gene family. Further online bioinformatics analysis revealed that miR-142-5p had potential binding sites that can bind circ_0004674 and the 3′UTR of MCL1 mRNA. Moreover, the expression and function of miR-142-5p were conversely correlated with circ_0004674 in vitro. RIP, pull-down, luciferase assay, and RNA FISH demonstrated that circ_0004674 could compete with MCL1 for miR-142-5p binding to counteract miR-142-5p-mediated repression of MCL1 at the post-transcriptional level. To sum up, our study sheds light on the critical role of the oncogenic circ_0004674/miR-142-5p/MCL1 axis in OS progression and chemoresistance, providing a novel potential target for OS therapy.
Similar content being viewed by others
Introduction
Osteosarcoma (OS), the most common primary malignant tumor of bone with peak incidence in children and adolescents, is highly invasive and tends to metastasize to the lung [1, 2]. Chemotherapy combined with complete surgical resection has been the standard treatment for most OS patients with a significantly improved 5-year disease-free survival rate to 70–80% [3]. But the survival rate of patients who suffer from chemoresistance or lung metastasis has declined to less than 20% [4, 5]. Therefore, it is essential to expand the molecular understanding of tumorigenesis and progression of OS to provide novel biomarkers or treatment targets for this refractory disease [6, 7].
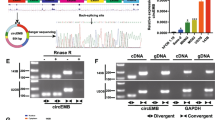
With the development of deep-sequencing technology, many novel noncoding RNAs, including circular RNAs (circRNAs), have been identified and found to act a pivotal part in diverse cellular physiological and pathological processes [8, 9]. Unlike its homologous linear transcript, the characteristic of circRNA is that it has a closed-loop structure without a 3′ polyadenylate tail or 5′ cap, making it highly tolerant to RNase R [10]. In addition, many reports have demonstrated that circRNAs can regulate the expression of numerous genes at the pre-, transcriptional, and post-transcriptional levels through interacting with DNA, RNA or protein [8, 11]. Lately, some circRNAs have been discovered to take part in the progression of OS, such as circMYO10 [RNase R treatment and Actinomycin D Total RNA of circ_0004674 and ADAM22 (10 μg) was incubated with RNase R (R0301, Geneseed, Guangzhou, China) at 37 °C for 60 min to analyze the RNase R resistance. Actinomycin D (2 mg/mL, 129935, Millipore, USA) was added to the cell culture medium, and the half-lives of circ_0004674 and ADAM22 were evaluated and analyzed. After treatment with actinomycin D or RNase R, the expression levels of circ_0004674 and ADAM22 were detected by qRT-PCR. MG63/DXR and KH-OS/DXR cells were transiently transfected with siRNAs (or miR-142-5p mimicss, miR-142-5p inhibitor, miR-NC) using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions after being seeded into 6-well plates overnight. Forty-eight hours after transfection, the cells were harvested to detect the knockout efficiency via qRT-PCR detection of the expression of circ_0004674 and ADAM22. Two different siRNAs against the junction site of circ_0004674 were designed and synthesized by GenePharma (Shanghai, China). The sequence of si-circ_0004674-1 was 5′-GTCGTGCTAAATCAATTCAGA-3′, that of si-circ_0004674-2 was 5′-GTGCTAAA TCAATTCAGATGT-3′ (si-circ_0004674-1 has the highest inhibition efficiency, and si-circ_0004674 mentioned in the article refers to si-circ_0004674-1) and the relative si-NC sequence was 5′-AAUUCUCCGAACGUGUCACGU-3′. Stably transfected MG63/DXR (or KH-OS/DXR) cells (5 × 103 cell/well) were seeded in 96-well plates, freshly prepared medium containing several final concentrations of doxorubicin (0, 2, 4, 8, 16, 32, and 64 μg/ml) was added to the wells with three replicate wells for each concentration. After incubation for another 48 h, cell viability was measured using Cell Counting Kit-8 (CCK-8, Do**do, Japan) according to the manufacturer’s instructions. The absorbance of each well was measured with a microplate reader set at 450 nm. For the cell cycle assay, after 48 h of transfection, the cells were digested with trypsin, and the supernatant was discarded. Subsequently, the cells were washed with PBS, centrifuged, and fixed in 75% ethanol at 4 °C overnight. After washing twice with PBS, the cells were stained with propidium iodide (PI) at room temperature in the dark for 15 min before analysis. For the cell apoptosis assay, after 48 h of transfection, doxorubicin (8 μg/ml) was added to the wells. After incubation for another 12 h, the Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Diego, USA) was used to stain cells according to the manufacturer’s protocol. The cell cycle and apoptosis were then analyzed by FACS scan flow cytometry using FlowJo software (BD Biosciences). For the invasion assays, a 24-well Transwell chamber with the upper chamber coated with Matrigel (354230, BD, USA) was used. A total of 5.0 × 104 stably transfected MG63/DXR (or KH-OS/DXR) cells in 200 µL of serum-free DMEM were seeded in the upper chamber, and 600 µl of medium containing 10% FBS was placed in the lower chamber. After incubation for 24 h, cells on the upper membrane surface were wiped off using a cotton swab, and the invading cells that had traversed the membrane were stained with crystal violet and counted. A total of stably transfected MG63/DXR (or KH-OS/DXR) cells (5 × 105 cell/well) were seeded in 6-well plates. Wounds were then created on a monolayer of cells using a sterile pipette tip. Cells were further cultured with a medium containing 1% FBS for 48 h. The healing wounds were photographed twice at 0 h and 48 h after scratching, and then, the cell migration rate was calculated. Female nude (BALB/c) mice (4 weeks old) were purchased. Mice were divided into two groups according to the completely randomized method (N = 6/group). All procedures for the mouse experiments were approved by the Animal Experimental Ethics Committee of Shanghai Tenth People’s Hospital. MG63/DXR cells stably expressing sh-circ_0004674 or sh-NC were propagated, and 5 × 106 (100 µl) cells were inoculated into the medullary cavity of the right proximal tibia of mice. Tumor growth was examined at the indicated time points, and tumor volumes were measured. After 7 weeks, the mice were killed, and tumors were removed and weighed. The activity of Ki-67 and caspase3 in the tumors was measured by immunochemistry (IHC). The sequence of circ_0004674 was obtained from Circbase (http://www.circbase. org/cgi-bin/simplesearch.cgi), and visual graphics of circ_0004674 were obtained from CircPrimer, a software which is for annotating circRNAs and determining the specificity of circRNA primers. CircInteractome (https://circinteractome.nia.nih.gov/) was used to analyze the target miRNA of circ_0004674. All the downregulated miRNAs in OS previously reported were collected from the database of dbDEMC (database of Differentially Expressed MiRNAs in human Cancers, https://www.picb. ac.cn/dbDEMC/index.html) and miRCancer (microRNA Cancer Association Database, http://mircancer.ecu.edu/). RIP experiments were performed in MG63/DXR cells using an RIP Kit (Millipore, Billerica, MA) following its manufacturer’s protocol. Cells were lysed and incubated with RIP buffer containing A/G magnetic beads conjugated with Ago2 antibody (Abcam) and normal anti-IgG (Millipore) as a negative control. Finally, immunoprecipitated RNA was isolated and purified for qRT-PCR to analyze the expression levels. Biotin-coupled probe pull-down assays were performed to determine the interaction between circ_0004674 and miR-142-5p. Briefly, the miR-142-5p-WT or miR-142-5p- Mut probe was synthesized and biotinylated by GenePharma (Shanghai, China). RNA pull-down assays were carried out using the Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, Waltham, MA) following its manufacturer’s protocols. Finally, the RNA binding protein complexes were washed and eluted for qPCR analysis of circ_0004674. MG63/DXR cells were seeded at 5 × 104 cells/well in 24-well plates and allowed to settle overnight. The next day, cells were co-transfected with pmirGLO-circ_0004674 (or MCL1)-WT or-MUT reporter plasmids and miR-142-5p mimics. Twenty-four hours after transfection, 20 µl of the protein supernatant was added to 50 µl of the firefly luciferase substrate and mixed thoroughly. Then, the relative luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) and normalized against Renilla luciferase activity. Cy3-labeled circ_0004674, Cy5-labeled miR-142-5p, and DAPI-labeled U6 probes were obtained from GenePharma (Shanghai, China). RNA FISH was performed using a fluorescent in situ hybridization kit according to the manufacturer’s protocol (Thermo Fisher). For western blotting, the total protein was extracted using RIPA lysis buffer from cells (P0013, Beyotime, CA). Then, the protein loading buffer was added to the protein and mixed, and the mixture was boiled for 10 min. Next, a 30-μg protein sample was added to each well, electrophoresed, and subsequently transferred to a NC membrane (Immobilon-P Transfer Membrane, EMD Millipore Corporation, MA). The membrane was blocked with 5% skim milk for 2 h and then incubated with the primary antibody overnight at 4 °C, followed by incubation with the corresponding secondary antibody for 2 h at room temperature. The protein bands were visualized using an Odyssey scanner (LI-COR Biosciences, Lincoln, NE, USA). ImageJ software was used for semiquantitative analysis. The primary antibodies were MCL1 and β-actin. All statistical analyses were performed using SPSS 22.0 software (IBM) and GraphPad Prism 7.0 (GraphPad Software Inc, San Diego, CA, USA) software. All the experiments in the current study were independently performed at least three times and data are presented as the means ± SEM. Student’s t test or one-way ANOVA was used to evaluate the differences between groups for continuous data. Chi-square test was used to compare the distribution of stage between high- and low-expression circ_0004674 level groups for categorical data. Overall survival was calculated by Kaplan–Meier survival analysis and compared using the log-rank test. The correlation of circ_0004674 and MCL1 mRNA expression level in OS tissues was determined by Spearman’s correlation. Then, p values of <0.05 were considered statistically significant.Plasmid construction and cell transfection
CCK-8 assay
Flow cytometry
Transwell assay
Wound healing assay
Xenograft tumor assay
Bioinformatics prediction
RNA immunoprecipitation (RIP)
Biotin-coupled probe pull-down assay
Dual-luciferase reporter assay
RNA fluorescence in situ hybridization (FISH)
Western blot
Statistical analysis
Data availability
All data generated or analyzed during this study are included in this published article.
References
Jafari F, Javdansirat S, Sanaie S, Naseri A, Shamekh A, Rostamzadeh D, et al. Osteosarcoma: a comprehensive review of management and treatment strategies. Ann Diagnostic Pathol. 2020;49:151654.
Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol: HO. 2010;8:705–18.
Yamamoto N, Tsuchiya H. Chemotherapy for osteosarcoma - where does it come from? What is it? Where is it going? Expert Opin Pharmacother. 2013;14:2183–93.
Ferrari S, Serra M. An update on chemotherapy for osteosarcoma. Expert Opin Pharmacother. 2015;16:2727–36.
Rastogi S, Aggarwal A, Tiwari A, Sharma V. Chemotherapy in nonmetastatic osteosarcoma: recent advances and implications for develo** countries. J Glob Oncol. 2018;4:1–5.
Zhang L, Chan MTV, Wu WKK, Lilienthal I, Herold N. Targeting molecular mechanisms underlying treatment efficacy and resistance in osteosarcoma: a review of current and future strategies. Cell Prolif. 2020;21:6885.
Czarnecka AM. Molecular biology of osteosarcoma. Cancers (Basel). 2020;12:2130.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91.
Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–42.
Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836.
Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic acids. 2019;16:118–29.
Chen J, Liu G, Wu Y, Ma J, Wu H, **e Z, et al. CircMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of β-catenin/LEF1 complex via effects on chromatin remodeling. Mol Cancer. 2019;18:150.
Zheng S, Qian Z, Jiang F, Ge D, Tang J, Chen H, et al. CircRNA LRP6 promotes the development of osteosarcoma via negatively regulating KLF2 and APC levels. Am J Transl Res. 2019;11:4126–38.
Ren C, Liu J, Zheng B, Yan P, Sun Y, Yue B. The circular RNA circ-ITCH acts as a tumour suppressor in osteosarcoma via regulating miR-22. Artif Cells Nanomed Biotechnol. 2019;47:3359–67.
Kun-Peng Z, **ao-Long M, Lei Z, Chun-Lin Z, Jian-** H, Tai-Cheng Z. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics. 2018;10:1327–46.
Zhu KP, Zhang CL, Ma XL, Hu JP, Cai T, Zhang L. Analyzing the Interactions of mRNAs and ncRNAs to Predict Competing Endogenous RNA Networks in Osteosarcoma Chemo-Resistance. Mol Ther: J Am Soc Gene Ther. 2019;27:518–30.
Kun-Peng Z, **ao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14:321–30.
Kun-Peng Z, Chun-Lin Z, Jian-** H, Lei Z. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int J Biol Sci. 2018;14:1513–20.
Cheng D, Li J, Zhang L, Hu L. miR-142-5p suppresses proliferation and promotes apoptosis of human osteosarcoma cell line, HOS, by targeting PLA2G16 through the ERK1/2 signaling pathway. Oncol Lett. 2019;17:1363–71.
Li X, Chen W, ** Y, Xue R, Su J, Mu Z, et al. miR-142-5p enhances cisplatin-induced apoptosis in ovarian cancer cells by targeting multiple anti-apoptotic genes. Biochemical Pharmacol. 2019;161:98–112.
Su J, Ruan S, Dai S, Mi J, Chen W, Jiang S. NF1 regulates apoptosis in ovarian cancer cells by targeting MCL1 via miR-142-5p. Pharmacogenomics. 2019;20:155–65.
Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R, et al. The emerging functions and roles of circular RNAs in cancer. EMBO J. 2018;414:301–9.
Wang Z, Deng M, Chen L, Wang W, Liu G, Liu D, et al. Circular RNA Circ-03955 Promotes Epithelial-Mesenchymal Transition in Osteosarcoma by Regulating miR-3662/Metadherin Pathway. Front Oncol. 2020;10:545460.
Jiang X, Chen D. Circular RNA hsa_circ_0000658 inhibits osteosarcoma cell proliferation and migration via the miR-1227/IRF2 axis. J Cell Mol Med. 2020;25:510–20.
Ji X, Shan L, Shen P, He M. Circular RNA circ_001621 promotes osteosarcoma cells proliferation and migration by sponging miR-578 and regulating VEGF expression. Cell Death Dis. 2020;11:18.
Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochimica Et. Biophysica Acta. 2016;1859:163–8.
Sang Y, Chen B, Song X, Li Y, Liang Y, Han D, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther: J Am Soc Gene Ther. 2019;27:1638–52.
Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol. Cancer. 2019;18:13.
Bian L, Zhi X, Ma L, Zhang J, Chen P, Sun S, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p/FOXO4 axis. Biochem Biophys Res Commun. 2018;505:346–52.
Zhu W, Wang JP, Meng QZ, Zhu F, Hao XF. MiR-142-5p reverses the resistance to gefitinib through targeting HOXD8 in lung cancer cells. Eur Rev Med Pharmacol Sci. 2020;24:4306–13.
Klümper T, Bruckmueller H, Diewock T, Kaehler M, Haenisch S, Pott C, et al. Expression differences of miR-142-5p between treatment-naïve chronic myeloid leukemia patients responding and non-responding to imatinib therapy suggest a link to oncogenic ABL2, SRI, cKIT and MCL1 signaling pathways critical for development of therapy resistance. Exp Hematol Oncol. 2020;9:26.
Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865–77.
Acknowledgements
This project was supported by Grant from the Program of Shanghai Sailing Talent Program (No.20YF1437700), Climbing Talents Program of Shanghai Tenth People’s Hospital (2021SYPDRC021), National Natural Science Foundation of China (No.81872174, 82072963), and Program of Shanghai Academic Research Leader (No.19XD1402900).
Author information
Authors and Affiliations
Contributions
Z.K.P. and M.X.L. carried out the molecular genetic studies. M.X.L. and H.J.P. carried out the tumor-bearing nude mice and IHC assays. Z.T.C. carried out the tissue samples collection and statistical analysis. Z.K.P. and Z.C.L. designed of the study and performed the statistical analysis. M.X.L. and Z.K.P. wrote the manuscript and Z.C.L. helped to correct it.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was conducted in accordance with the declaration of Helsinki. This study was conducted with approval from the Ethics Committee of Shanghai Tenth People’s Hospital. Written informed consent was obtained from all participants.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ma, XL., Zhan, TC., Hu, JP. et al. Doxorubicin-induced novel circRNA_0004674 facilitates osteosarcoma progression and chemoresistance by upregulating MCL1 through miR-142-5p. Cell Death Discov. 7, 309 (2021). https://doi.org/10.1038/s41420-021-00694-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-021-00694-8
- Springer Nature Limited