Abstract
Cancer cells undergo metabolic reprogramming in response to hostile microenvironments, such as energy stress; however, the underlying mechanisms remain largely unclear. It is also unknown whether energy stress-responsive circular RNA (circRNA) is involved in the regulation of glucose metabolism. Here we report that circDDX21 is upregulated in response to glucose deprivation by the transcription factor c-Myc. Functionally, circDDX21 is shown to promote glycolysis by increasing PGAM1 expression. Mechanistically, circDDX21 interacts with the RNA binding protein PABPC1, disrupting its association with the ubiquitin E3 ligase MKRN3. This disassociation attenuates MKRN3-mediated PABPC1 ubiquitination and enhances the binding of PABPC1 to PGAM1 mRNA, thereby leading to PGAM1 mRNA stabilization. The ability of the circDDX21-PGAM1 axis to promote hepatocellular carcinogenesis is validated in a xenograft mouse model. Additionally, in clinical hepatocellular carcinoma tissues, there is a positive correlation between circDDX21 and PGAM1 expression. These findings establish circDDX21 as an important regulator of glycolysis and suggest circDDX21 as a potential therapeutic target for hepatocellular carcinoma.
Similar content being viewed by others
Introduction
Metabolic reprogramming is a crucial hallmark of cancer cells [1]. In comparison to their healthy counterparts, cancer cells exhibit an aberrant alteration in glucose metabolism, known as the Warburg effect. This effect is characterized by enhanced glycolysis and reduced oxidative phosphorylation, even in the presence of sufficient oxygen [2]. The Warburg effect, along with increased glucose uptake, not only enables cancer cells to meet their high energetic and biosynthetic demands but also creates an acidic environment, providing a growth advantage for tumors [3, 4]. However, during rapid tumor growth, nutrients such as glucose are often limited, which requires cancer cells to reprogram their metabolism for survival and proliferation [5, 6].
It has been well established that glycolysis plays a pivotal role in both cancer initiation and progression [7, 8]. The mechanisms responsible for increased glycolysis in cancer cells include upregulated expression and enhanced activity of several glycolytic enzymes [9, 10]. Among these glycolytic enzymes, phosphoglycerate mutase 1 (PGAM1) is a rate-limiting enzyme that catalyzes the reversible conversion between 3-phosphoglycerate (3-PG) and 2-phosphoglycerate (2-PG) [11]. PGAM1 is frequently upregulated in various human cancers, including breast cancer, lung cancer, and hepatocellular carcinoma [12,13,14]. Overexpression of PGAM1 promotes cancer progression, whereas inhibition of PGAM1 using shRNAs or inhibitors effectively suppresses cancer progression [15,16,17,18,19]. Given the importance of PGAM1 in cancer progression, it is not surprising that PGAM1 is intricately regulated by multiple factors at the transcriptional, translational, and post-translational levels [20,21,22,48]. Subsequent in vitro binding assay showed that unlike wild-type circDDX21, mutant circDDX21 (ΔPABPC1 BS) with mutation of the predicted PABPC1-binding site did not exhibit obvious PABPC1-binding ability (Supplementary Fig. 4J, K). These data support that PABPC1 is a binding partner for circDDX21.
We next evaluated whether circDDX21 positively regulates PGAM1 mRNA stability via PABPC1. The results showed that the decreased PGAM1 mRNA stability caused by circDDX21 knockdown could be significantly restored by overexpression of PABPC1. (Fig. 4G). In accordance, the decreased mRNA and protein levels of PGAM1 resulting from circDDX21 knockdown could be markedly rescued by ectopically expressed PABPC1 (Fig. 4H, I, Supplementary Fig. 4L, M). In addition, circDDX21 overexpression was shown to increase the mRNA and protein levels of PGAM1 in control cells, but not in PABPC1 knockdown cells (Fig. 4J, K, Supplementary Fig. 4N, O). Moreover, compared to wild-type circDDX21, mutant circDDX21 (ΔPABPC1 BS) without PABPC1-binding ability did not exhibit noticeable effects on PGAM1 mRNA and protein levels (Fig. 4L, M). These data indicate that the regulatory effect of circDDX21 on PGAM1 expression is dependent on PABPC1. Considering the 3’-UTR is critical for controlling mRNA stability, we cloned the PGAM1 3’-UTR into the psiCHECK2 luciferase reporter construct (Supplementary Fig. 4P). As expected, the reporter activity from the psiCHECK2-PGAM1 3’-UTR was reduced by circDDX21 knockdown and increased by circDDX21 overexpression (Fig. 4N, O). The inhibitory effect of circDDX21 knockdown on PGAM1 3’-UTR reporter activity could be reversed by PABPC1 overexpression (Fig. 4N). In addition, circDDX21 failed to increase PGAM1 3’-UTR reporter activity when PABPC1 was knocked down (Fig. 4O). These findings indicate that the stabilizing effect of circDDX21 on PGAM1 mRNA depends on PABPC1. These results also raised the question of whether circDDX21 could facilitate the binding of PABPC1 to PGAM1 mRNA, thereby enhancing PGAM1 mRNA stability. To address this, an RNA immunoprecipitation assay was performed. The results showed that knockdown of circDDX21 attenuated, whereas overexpression of circDDX21 increased, the binding of PABPC1 to PGAM1 mRNA (Fig. 4P, Q). Taken together, these data suggest that circDDX21 cooperates with PABPC1 to promote PGAM1 mRNA stabilization.
circDDX21 increases the binding of PABPC1 to PGAM1 mRNA by inhibiting MKRN3-mediated PABPC1 ubiquitination
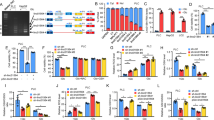
We next sought to investigate how circDDX21 enhances the binding of PABPC1 to PGAM1 mRNA. It has been previously shown that MKRN3-mediated ubiquitination attenuates the binding of PABPC1 to its target mRNA [49, 50]. We therefore asked whether circDDX21 could regulate PABPC1 ubiquitination, thereby affecting the interaction between PABPC1 and PGAM1 mRNA. By performing an in vivo ubiquitination assay, we showed that overexpression of wild-type circDDX21, but not PABPC1 binding-defective mutant of circDDX21 (ΔPABPC1 BS), greatly decreased PABPC1 ubiquitination (Fig. 5A, Supplementary Fig. 5A). Conversely, knockdown of circDDX21 strongly increased PABPC1 ubiquitination (Fig. 5B). To determine whether the inhibitory effect of circDDX21 on PABPC1 ubiquitination is dependent on MKRN3, we performed rescue experiments. In agreement with previous reports [49, 50], ubiquitination of endogenous PABPC1 was induced by MKRN3 overexpression and reduced by MKRN3 knockdown (Fig. 5C, D), reinforcing the importance of MKRN3 in controlling PABPC1 ubiquitination. Moreover, the circDDX21-mediated suppression of PABPC1 ubiquitination could be reversed by ectopic expression of MKRN3 (Fig. 5C), while knockdown of MKRN3 completely abolished the enhanced PABPC1 ubiquitination caused by circDDX21 knockdown (Fig. 5D), indicating that circDDX21 suppresses MKRN3-mediated PABPC1 ubiquitination.
A Lysates from HepG2 cells transduced with lentiviruses expressing empty vector (EV) or circDDX21 were subjected to an in vivo ubiquitination assay using anti-PABPC1 antibody. B Lysates from HepG2 cells transduced with lentiviruses expressing control shRNA or circDDX21 shRNA were subjected to an in vivo ubiquitination assay using an anti-PABPC1 antibody. C HepG2 cells were infected with lentiviruses expressing circDDX21 and Flag-MKRN3 in the indicated combination, followed by an in vivo ubiquitination assay using an anti-PABPC1 antibody. D HepG2 cells were infected with lentiviruses expressing circDDX21 shRNA and MKRN3 sgRNA in the indicated combination, followed by an in vivo ubiquitination assay using an anti-PABPC1 antibody. E HepG2 cells were infected with lentiviruses expressing circDDX21 and HA-MKRN3 in the indicated combination, followed by an RNA immunoprecipitation assay using an anti-PABPC1 antibody. F HepG2 cells were infected with lentiviruses expressing circDDX21 shRNA and MKRN3 sgRNA in the indicated combination, followed by an RNA immunoprecipitation assay using an anti-PABPC1 antibody. G HepG2 cells were infected with lentiviruses expressing circDDX21 and Flag-MKRN3 in the indicated combination, followed by real-time RT-PCR analysis of PGAM1 mRNA levels. Data shown are mean ± SD (n = 3). **p < 0.01, ***p < 0.001. H HepG2 cells were infected with lentiviruses expressing circDDX21 and Flag-MKRN3 in the indicated combination, followed by western blot analysis of PGAM1 protein levels. I HepG2 cells were infected with lentiviruses expressing circDDX21 shRNA and MKRN3 sgRNA in the indicated combination, followed by real-time RT-PCR analysis of PGAM1 mRNA levels. Data shown are mean ± SD (n = 3). *p < 0.05, **p < 0.01. J HepG2 cells were infected with lentiviruses expressing circDDX21 shRNA and MKRN3 sgRNA in the indicated combination, followed by western blot analysis of PGAM1 protein levels. K HepG2 cells were infected with lentiviruses expressing circDDX21 and Flag-MKRN3 in the indicated combination, followed by an immunoprecipitation assay using anti-Flag antibody. L HepG2 cells were infected with lentiviruses expressing circDDX21 shRNA and Flag-MKRN3 in the indicated combination, followed by an immunoprecipitation assay using anti-Flag antibody.
Consistent with the role of MKRN3 in circDDX21-regulated PABPC1 ubiquitination, the enhancing effect of circDDX21 on the PABPC1-PGAM1 mRNA interaction was strongly minimized by MKRN3 overexpression (Fig. 5E). In addition, the decreased PABPC1-PGAM1 mRNA interaction resulting from circDDX21 knockdown could be substantially rescued by concurrent knockdown of MKRN3 (Fig. 5F). Collectively, these data suggest that circDDX21 enhances the binding of PABPC1 to PAGM1 mRNA by suppressing MKRN3-mediated PABPC1 ubiquitination. In accordance, circDDX21 overexpression was shown to induce the mRNA and protein expression of PGAM1 in control cells, but not in MKRN3-overexpressing cells (Fig. 5G, H). Moreover, the reduction in mRNA and protein levels of PGAM1 caused by circDDX21 knockdown could be greatly recovered by concurrent knockdown of MKRN3 (Fig. 5I, J).
We next explored how circDDX21 suppresses MKRN3-mediated PABPC1 ubiquitination. The immunoprecipitation assay revealed that the interaction between MKRN3 and PABPC1 was compromised by circDDX21 overexpression (Fig. 5K). In contrast, circDDX21 knockdown enhanced the MKRN3-PABPC1 interaction (Fig. 5L). By performing map** experiments, circDDX21 was shown to associate with both the central region (aa 181-380) and the C-terminal region (aa 381-636) of PABPC1 (Supplementary Fig. 5B, C). This C-terminal region (aa 381-636) of PABPC1 has been previously reported to mediate the interaction with MKRN3 [49]. Therefore, our data imply that circDDX21 competes with MKRN3 for binding to the C-terminal region (aa 381-636) of PABPC1, thereby disrupting the MKRN3-PABPC1 association and suppressing MKRN3-mediated PABPC1 ubiquitination.
Biological implication of circDDX21 in hepatocellular carcinogenesis
Given the promoting effect of circDDX21 on glycolysis in hepatocellular carcinoma cells, as demonstrated above, we asked whether circDDX21 could facilitate hepatocellular carcinogenesis. We first examined the effect of circDDX21 on the proliferation of hepatocellular carcinoma HepG2 cells. Knockdown of circDDX21 in HepG2 cells resulted in a dramatic decrease in proliferation and colony numbers (Fig. 6A, B). Conversely, overexpression of circDDX21 led to a strong increase in cell proliferation and colony formation (Fig. 6C, D). The inhibitory effects of circDDX21 knockdown on cell proliferation and colony formation were markedly reversed by ectopically expressed PGAM1 (Fig. 6A, B). In addition, circDDX21 no longer increased cell proliferation and colony formation when PGAM1 was knocked down (Fig. 6C, D). These data suggest that circDDX21 promotes hepatocellular carcinoma cell proliferation via PGAM1.
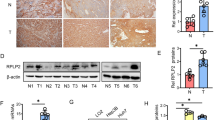
A Growth curves of HepG2 cells expressing control, circDDX21 shRNA, PGAM1, or both circDDX21 shRNA and PGAM1. Data shown are mean ± SD (n = 3). *p < 0.05, ***p < 0.001. B Colonies of HepG2 cells expressing control, circDDX21 shRNA, PGAM1, or both circDDX21 shRNA and PGAM1 were stained with crystal violet after 14 days of incubation. Data shown are mean ± SD (n = 3). **p < 0.01, ***p < 0.001. C Growth curves of HepG2 cells expressing control, circDDX21, PGAM1 shRNA, or both circDDX21 and PGAM1 shRNA. Data shown are mean ± SD (n = 3). **p < 0.01, ***p < 0.001, ns, no significance. D Colonies of HepG2 cells expressing control, circDDX21, PGAM1 shRNA, or both circDDX21 and PGAM1 shRNA were stained with crystal violet after 14 days of incubation. Data shown are mean ± SD (n = 3). *p < 0.05, ***p < 0.001, ns, no significance. E Growth curves of HepG2 cells expressing control, circDDX21 shRNA, PABPC1, or both circDDX21 shRNA and PABPC1. Data shown are mean ± SD (n = 3). *p < 0.05, ***p < 0.001. F Colonies of HepG2 cells expressing control, circDDX21 shRNA, PABPC1, or both circDDX21 shRNA and PABPC1 were stained with crystal violet after 14 days of incubation. Data shown are mean ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001. G Growth curves of HepG2 cells expressing control, circDDX21, PABPC1 shRNA, or both circDDX21 and PABPC1 shRNA. Data shown are mean ± SD (n = 3). *p < 0.05, ***p < 0.001, ns, no significance. H Colonies of HepG2 cells expressing control, circDDX21, PABPC1 shRNA, or both circDDX21 and PABPC1 shRNA were stained with crystal violet after 14 days of incubation. Data shown are mean ± SD (n = 3). ***p < 0.001, ns, no significance. A total of 3 × 106 HepG2 cells expressing control, circDDX21 shRNA, PGAM1, or both circDDX21 shRNA and PGAM1 were individually injected into nude mice (n = 6 for each group). I Xenograft tumors were taken 24 days after injection. J Excised tumors were weighed. **p < 0.01, ***p < 0.001. K Tumor sizes were measured at the indicated time points. ***p < 0.001. L RNA and protein extracts from the excised xenografts were analyzed by RT-PCR and western blotting, respectively. M Real-time RT-CR analysis of circDDX21 levels among 25 pairs of matched hepatocellular carcinoma and adjacent normal tissue. Data shown are mean ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, ns., no significance. The dotted line indicates a fold change of 2. N Correlation analyses conducted between circDDX21 and PGAM1 mRNA expression in hepatocellular carcinoma samples (n = 51).
We next evaluated the functional role of PABPC1 in circDDX21-promoted cell proliferation. Ectopic expression of PABPC1 greatly rescued circDDX21 knockdown-induced decreases in cell proliferation and colony formation (Fig. 6E, F). In addition, circDDX21 overexpression consistently increased cell proliferation and colony formation in control cells, but not in PABPC1 knockdown cells (Fig. 6G, H), indicating that PABPC1 plays an important role in mediating the promoting effect of circDDX21 on cell proliferation. To determine whether circDDX21 promotes cell proliferation via the PABPC1-PGAM1 axis, we utilized PABPC1 binding-defective mutant of circDDX21 (ΔPABPC1 BS), which lost the ability to induce PGAM1 expression (Fig. 4L, M, Supplementary Fig. 4J, K). Unlike wild-type circDDX21, this mutant circDDX21 (ΔPABPC1 BS) failed to promote cell proliferation and colony formation (Supplementary Fig. 6A, B). In accordance, mutant circDDX21 (ΔPABPC1 BS) exhibited no obvious effect on the glycolytic rate compared to wild-type circDDX21 (Supplementary Fig. 6C). These data imply an important role of the PABPC1-PGAM1 axis in circDDX21-accelerated cell proliferation.
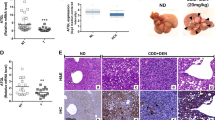
By using a xenograft mouse model, knockdown of circDDX21 was shown to significantly inhibit in vivo xenograft tumor growth of HepG2 cells (Fig. 6I–L). In contrast, overexpression of circDDX21 in HepG2 cells substantially enhanced in vivo xenograft tumor growth (Supplementary Fig. 6D–G). CircDDX21 knockdown-reduced xenograft tumor growth was markedly recovered by PGAM1 overexpression (Fig. 6I-L). The enhancing effect of circDDX21 on xenograft tumor growth was completely diminished by PGAM1 knockdown (Supplementary Fig. 6D–G). These data suggest that circDDX21 facilitates in vivo hepatocellular carcinoma cell growth via PGAM1. To further validate the clinical implication of circDDX21 in hepatocellular carcinogenesis, we examined the expression levels of circDDX21 in hepatocellular carcinoma tissues and matched adjacent normal tissues. Compared to the normal tissue samples, over 70% (18 of 25) of hepatocellular carcinoma (HCC) tissue samples exhibited elevated expression of circDDX21 (Fig. 6M). Furthermore, circDDX21 expression was positively correlated with PGAM1 expression in all the examined hepatocellular carcinoma tissue samples (n = 51) (Fig. 6N), indicating the biological importance of the circDDX21-PGAM1 axis in hepatocellular carcinogenesis. Taken together, these data strongly support that circDDX21 functions as an oncogenic circRNA in hepatocellular carcinoma.
Discussion
Cancer cells actively rewire their metabolism in response to energy stress conditions in the tumor microenvironment [51, 52]. Increased glycolysis has been recognized as one of the major hallmarks of cancer cells [2]. However, it remains unknown whether energy stress-responsive circRNA plays a regulatory role in glycolysis. In this study, we report that circDDX21, as a glucose deprivation-induced circRNA, promotes glycolysis by increasing the expression of the glycolytic enzyme PGAM1. Therefore, circDDX21 may represent an important molecule that links glycolysis to the glucose-deprived microenvironment of tumors.
As a master transcription factor, c-Myc has been implicated in the regulation of cell metabolism by modulating target gene expression under both non-stressed and stressed conditions [53]. For instance, c-Myc is activated under energy stress conditions, which in turn upregulates the expression of key genes in the serine synthesis pathway and results in increased serine synthesis [54]. Here, we present evidence showing that c-Myc is responsible for the increased expression of circDDX21 in response to glucose deprivation. This finding, together with the critical role of circDDX21 in promoting glycolysis, suggests that circRNAs may be an important class of molecules that connect c-Myc to metabolic reprogramming in cancer cells.
The glycolytic enzyme PGAM1 plays a critical role in cancer metabolism by coordinating glycolysis and biosynthesis to promote rapid tumor growth [16]. Given the importance of PGAM1 in regulating cancer metabolism, the expression and activity of PGAM1 are not surprisingly controlled at different levels. For instance, PGAM1 is transcriptionally regulated by the transcription factors p53 and HIF-1α [20, 24]. PGAM1 is also subjected to post-translational modifications such as acetylation and phosphorylation, which affect its enzymatic activity [22, 23]. Moreover, the lncRNAs NEAT1 and glycoLINC can act as scaffold molecules for PGAM1 to interact with other glycolytic enzymes, thereby ensuring the efficiency of glycolysis [55, 56]. Here, we show that, correlating with its predominant cytoplasmic localization, circDDX21 is able to promote PGAM1 mRNA stability, indicating that circDDX21 is an important factor that finely controls PGAM1 expression. These findings also highlight the complexity of PGAM1 regulation.
CircRNAs have been shown to regulate gene expression via multiple mechanisms. One of the well-accepted mechanisms is circRNA-mediated gene regulation through interaction with their target proteins [57]. Here, we show that circDDX21 cooperates with the RNA binding protein PABPC1 to stabilize PGAM1 mRNA. PABPC1 belongs to the PABP family of proteins that bind poly-A or AU-rich sequences in mRNAs [58]. PABPC1 is able to regulate different aspects of RNA metabolism, including mRNA stability and translation [59,60,61]. CircDDX21 is identified as a new PABPC1 binding partner. The putative PABPC1-binding site within circDDX21 appears to be necessary for the interaction with PABPC1. We also show that by binding to PABPC1, circDDX21 enhances the association between PABPC1 and PGAM1 mRNA, thereby stabilizing PGAM1 mRNA. It has been previously reported that PABPC1 undergoes MKRN3-mediated ubiquitination, which inhibits the binding of PABPC1 to its target mRNA [49, 50]. Intriguingly, circDDX21 appears to compete with MKRN3 for binding to PABPC1 and suppresses MKRN3-mediated PABPC1 ubiquitination, thereafter increasing the interaction of PABPC1 with PGAM1 mRNA. These findings suggest an important role of the MKRN3-PABPC1 axis in mediating the promoting effect of circDDX21 on PGAM1 mRNA stability. It has been recently reported that circDDX21 functions as a sponge for miR-1264 to regulate QKI expression in triple-negative breast cancer [62]. Therefore, it would be interesting to investigate whether circDDX21 could also act as a miRNA sponge to regulate PGAM1 mRNA stability in the future.
Overexpression of PGAM1 has been found in various human cancers, and increased expression of PGAM1 is associated with poor prognosis in cancer patients [19, 63]. In this study, we show that consistent with the enhancing effect of circDDX21 on PGAM1 expression, circDDX21 is able to promote both in vitro hepatocellular carcinoma cell proliferation and in vivo xenograft tumor growth via PGAM1. In addition, circDDX21 is highly expressed in clinical hepatocellular carcinoma tissues compared to normal tissues. The expression of circDDX21 and PGAM1 are also positively correlated in hepatocellular carcinoma. These data support that circDDX21 functions as an oncogenic circRNA in hepatocellular carcinoma. Unlike this oncogenic function in hepatocellular carcinoma, circDDX21 seems to play a tumor-suppressive role in triple-negative breast cancer [62], indicating that circDDX21 may function in vivo as an oncogene or tumor suppressor in different cancer types. Taken together, our study demonstrates an important role of circDDX21 in promoting glycolysis and hepatocellular carcinogenesis, and suggests that circDDX21 may represent a potential therapeutic target for hepatocellular carcinoma.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
References
Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46.
Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:618–618.
Heiden MGV, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33.
Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:287–287.
Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–65.
Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–9.
Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–9.
Abdel-Wahab AF, Mahmoud W, Al-Harizy RM. Targeting glucose metabolism to suppress cancer progression: prospective of anti-glycolytic cancer therapy. Pharmacol Res. 2019;150:104511.
Nowak N, Kulma A, Gutowicz J. Up-regulation of key glycolysis proteins in cancer development. Open Life Sci. 2018;13:569–81.
Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–98.
Fothergillgilmore LA, Watson HC. The phosphoglycerate mutases. Adv Enzymol Ramb. 1989;62:227–313.
Durany N, Joseph J, Jimenez OM, Climent F, Fernández PL, Rivera F, et al. Phosphoglycerate mutase, 2,3 bisphosphoglycerate phosphatase, creatine kinase and enolase activity and isoenzymes in breast carcinoma. Br J Cancer. 2000;82:20–27.
Ren FL, Wu H, Lei YL, Zhang HY, Liu R, Zhao Y, et al. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol Cancer. 2010;9:81.
Durany N, Joseph J, Campo E, Molina R, Carreras J. Phosphoglycerate mutase, 2,3-bisphosphoglycerate phosphatase and enolase activity and isoenzymes in lung, colon and liver carcinomas. Br J Cancer. 1997;75:969–77.
Evans MJ, Saghatelian A, Sorensen EJ, Cravatt BF. Target discovery in small-molecule cell-based screens by proteome reactivity profiling. Nat Biotechnol. 2005;23:1303–7.
Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600.
Wen CL, Huang K, Jiang LL, Lu XX, Dai YT, Shi MM, et al. An allosteric PGAM1 inhibitor effectively suppresses pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA. 2019;116:23264–73.
Huang K, Liang Q, Zhou Y, Jiang LL, Gu WM, Luo MY, et al. A novel allosteric inhibitor of phosphoglycerate mutase 1 suppresses growth and metastasis of non-small-cell lung cancer. Cell Metab. 2019;30:1107–19.
Yang GJ, Tao F, Zhong HJ, Yang C, Chen J. Targeting PGAM1 in cancer: an emerging therapeutic opportunity. Eur J Med Chem. 2022;244:114798.
Sun Q, Li SZ, Wang YN, Peng HY, Zhang XY, Zheng Y, et al. Phosphoglyceric acid mutase-1 contributes to oncogenic mTOR-mediated tumor growth and confers non-small cell lung cancer patients with poor prognosis. Cell Death Differ. 2018;25:1160–73.
Heiden MGV, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–9.
Hallows WC, Yu W, Denu JM. Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. J Biol Chem. 2012;287:3850–8.
Hitosugi T, Zhou L, Fan J, Elf S, Zhang L, **e JX, et al. Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation. Nat Commun. 2013;4:1790.
Corcoran CA, Huang Y, Sheikh MS. The regulation of energy generating metabolic pathways by p53. Cancer Biol Ther. 2006;5:1610–3.
Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22:1760–74.
Wu WY, Ji PF, Zhao FQ. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020;21:101.
Deng SJ, Chen HY, Zeng Z, Deng SC, Zhu S, Ye Z, et al. Nutrient stress-dysregulated antisense lncRNA GLS-AS impairs GLS-mediated metabolism and represses pancreatic cancer progression. Cancer Res. 2019;79:1398–412.
Liu XW, **ao ZD, Han L, Zhang JX, Lee SW, Wang WQ, et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 2016;18:431–42.
Ma ML, Xu HX, Liu G, Wu J, Li CH, Wang XX, et al. Metabolism-induced tumor activator 1 (MITA1), an energy stress-inducible long noncoding RNA, promotes hepatocellular carcinoma metastasis. Hepatology. 2019;70:215–30.
**ao ZD, Han L, Lee H, Zhuang L, Zhang YL, Baddour J, et al. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat Commun. 2017;8:783.
Tang JY, Yan TT, Bao YJ, Shen CQ, Yu CY, Zhu XQ, et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat Commun. 2019;10:3499.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91.
Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836.
Liu CX, Chen LL. Circular RNAs: characterization, cellular roles, and applications. Cell. 2022;185:2016–34.
Yang L, Wilusz JE, Chen LL. Biogenesis and regulatory roles of circular RNAs. Annu Rev Cell Dev Biol. 2022;38:263–89.
Li J, Hu ZQ, Yu SY, Mao L, Zhou ZJ, Wang PC, et al. CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res. 2022;82:1055–69.
Cai ZR, Hu Y, Liao K, Li H, Chen DL, Ju HQ. Circular RNAs: emerging regulators of glucose metabolism in cancer. Cancer Lett. 2023;552:215978.
Li Q, Pan XX, Zhu DM, Deng ZM, Jiang RQ, Wang XH. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70:1298–316.
Li HH, Yang F, Hu AP, Wang XJ, Fang EH, Chen YJ, et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol Med. 2019;11:e10835.
Zhou JY, Zhang SZ, Chen ZM, He ZF, Xu Y, Li ZJ. CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis. 2019;10:885.
Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206.
Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther-Nucl Acids. 2019;16:118–29.
Chen L, Huang C, Shan G. Circular RNAs in physiology and non-immunological diseases. Trends Biochem Sci. 2022;47:250–64.
Li QD, Wang YC, Wu S, Zhou Z, Ding XJ, Shi RH, et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 2019;30:157–73.
Dong Y, Tu RF, Liu HD, Qing GL. Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat. Signal Transduct Tar. 2020;5:124.
Meliala ITS, Hosea R, Kasim V, Wu SR. The biological implications of Yin Yang 1 in the hallmarks of cancer. Theranostics. 2020;10:4183–4200.
Hirotsu Y, Higashi C, Fukutomi T, Katsuoka F, Tsujita T, Yagishita Y, et al. Transcription factor NF-E2-related factor 1 impairs glucose metabolism in mice. Genes Cells. 2014;19:650–65.
Paz I, Kosti I, Ares M, Cline M, Mandel-Gutfreund Y. RBPmap: a web server for map** binding sites of RNA-binding proteins. Nucleic Acids Res. 2014;42:W361–W367.
Li CAY, Han TT, Li QR, Zhang MH, Guo R, Yang Y, et al. MKRN3-mediated ubiquitination of Poly(A)-binding proteins modulates the stability and translation of mRNA in mammalian puberty. Nucleic Acids Res. 2021;49:3796–813.
Li K, Zheng XF, Tang H, Zang YS, Zeng CL, Liu XX, et al. E3 ligase MKRN3 is a tumor suppressor regulating PABPC1 ubiquitination in non-small cell lung cancer. J Exp Med. 2021;218:e20210151.
Pavlova NN, Zhu JJ, Thompson CB. The hallmarks of cancer metabolism: still emerging. Cell Metab. 2022;34:355–77.
Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669–80.
Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–39.
Sun LC, Song LB, Wan QF, Wu GW, Li XH, Wang YH, et al. cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 2015;25:429–44.
Park MK, Zhang L, Min KW, Cho JH, Yeh CC, Moon H, et al. NEAT1 is essential for metabolic changes that promote breast cancer growth and metastasis. Cell Metab. 2021;33:2380–97.
Zhu YM, ** L, Shi RH, Li JM, Wang Y, Zhang L, et al. The long noncoding RNA glycoLINC assembles a lower glycolytic metabolon to promote glycolysis. Mol Cell. 2022;82:542–54.
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19:172.
Wigington CP, Williams KR, Meers MP, Bassell GJ, Corbett AH. Poly(A) RNA-binding proteins and polyadenosine RNA: new members and novel functions. Wires Rna. 2014;5:601–22.
Burgess HM, Gray NK. mRNA-specific regulation of translation by poly(A)-binding proteins. Biochem Soc Trans. 2010;38:1517–22.
Brook M, Gray NK. The role of mammalian poly(A)-binding proteins in co-ordinating mRNA turnover. Biochem Soc Trans. 2012;40:856–64.
Qi Y, Wang M, Jiang Q. PABPC1–mRNA stability, protein translation and tumorigenesis. Front Oncol. 2022;12:1025291.
Huang RH, Yang Z, Liu Q, Liu B, Ding XY, Wang Z. CircRNA DDX21 acts as a prognostic factor and sponge of miR-1264/QKI axis to weaken the progression of triple-negative breast cancer. Clin Transl Med. 2022;12:e768.
Chen GA, Gharib TG, Wang H, Huang CC, Kuick R, Thomas DG, et al. Protein profiles associated with survival in lung adenocarcinoma. Proc Natl Acad Sci USA. 2003;100:13537–42.
Acknowledgements
We thank Dr. **ng Chang (Westlake University, China) for providing us PABPC1 expressing plasmid. This work was supported by grants from the Ministry of Science and Technology of China (2019YFA0802600), National Natural Science Foundation of China (32270811 and 82203562), Center for Advanced Interdisciplinary Science and Biomedicine of IHM (QYPY20220006), and the Fundamental Research Funds For Central Universities (YD9100002012).
Author information
Authors and Affiliations
Contributions
JL, YY, YM, and FW conceived and designed the project. JL, YY, GZ, DF, and KL performed all the experiments. JL, YY, GZ, DF, KL, YM, and FW analyzed the data. JL, YM, and FW wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Barak Rotblat
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Luo, J., Yang, Y., Zhang, G. et al. Energy stress-induced circDDX21 promotes glycolysis and facilitates hepatocellular carcinogenesis. Cell Death Dis 15, 354 (2024). https://doi.org/10.1038/s41419-024-06743-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-024-06743-1
- Springer Nature Limited