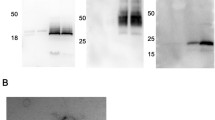
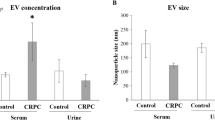
Abstract
Extracellular vesicles (EVs) and their cargo represent an intriguing source of cancer biomarkers for develo** robust and sensitive molecular tests by liquid biopsy. Prostate cancer (PCa) is still one of the most frequent and deadly tumor in men and analysis of EVs from biological fluids of PCa patients has proven the feasibility and the unprecedented potential of such an approach. Here, we exploited an antibody-based proteomic technology, i.e. the Reverse-Phase Protein microArrays (RPPA), to measure key antigens and activated signaling in EVs isolated from sera of PCa patients. Notably, we found tumor-specific protein profiles associated with clinical settings as well as candidate markers for EV-based tumor diagnosis. Among others, PD-L1, ERG, Integrin-β5, Survivin, TGF-β, phosphorylated-TSC2 as well as partners of the MAP-kinase and mTOR pathways emerged as differentially expressed endpoints in tumor-derived EVs. In addition, the retrospective analysis of EVs from a 15-year follow-up cohort generated a protein signature with prognostic significance. Our results confirm that serum-derived EV cargo may be exploited to improve the current diagnostic procedures while providing potential prognostic and predictive information. The approach proposed here has been already applied to tumor entities other than PCa, thus proving its value in translational medicine and paving the way to innovative, clinically meaningful tools.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) is still the second cause of cancer-related male deaths in highly developed countries [1]. A significant fraction of PCa patients arrives at diagnosis with advanced forms, while others retain indolent tumors which will never progress into aggressive stages [2, 3]. Therefore, an accurate, early diagnosis is likely to improve the outcome and the quality of life of PCa patients while reducing the over-treatment [4].
Extracellular vesicles (EVs) are membrane-enclosed bodies in the nano- to micro-meter scale that are secreted by nearly all cells and shuttle their biological content as a means of cell-to-cell communication [5, 6]. Tumor cells are now recognized to release more EVs than their normal counterpart and tumor-derived EVs can be easily isolated from bodily fluids [7,8,9,10], thus offering an exquisite source in terms of biomarkers and, mechanistically, of cancer treatment strategies [11,12,13]. The EV sub-population in the range of 30–150 nm in diameter is referred to as exosomes and has been shown to actively transport DNA, proteins, long and small RNAs [11, 14] as well as small peptides, such as prions [15]. Different from other vesicles, which are generated by random shedding mechanisms or from dying cells by discharge, exosomes drive intra- and inter-tissue cross-talk [16,17,18], are involved in physiological tissue homeostasis and immune system regulation [11] and in processes [12, 19, 20] that are often aberrant in tumors [7]. In this regard, PCa is characterized by multiple genomic lesions [Luminex assay 100 μg of EVs were lysed in 50 μl of standard RIPA buffer [(20 mM Tris-HCl pH7.2150 mM NaCl;1% NP40 (Igepal CA-630); Distilled water to volume; Proteases-inhibitors)] and diluted 1:4 in PBS for the analysis. 100 μg of parental EVs were left, non-lysed (SN), in 50 μl in the buffer (PBS) of the last step of Ultracentrifugation and was directly analyzed by Luminex. Cytokine/chemokine quantification in EV extracts and in EV(SN) was achieved by xMAP technology through a Luminex platform (Bio-Rad Laboratories, Hercules, CA, USA) equipped with a magnetic washer workstation according to the manufacturer’s protocol. RIPA (dilute 1:4 in PBS) and PBS buffer were used as background controls. Samples were analyzed using a human magnetic Luminex assay (R&D Systems, Minneapolis, MN, USA). Brain-Derived Neutrophil Factor (BDNF), CCL11, Fibroblast Growth Factor 13 (FGF-13), IL-5, IL-4, IL-23, IL-6, MMP-2 (membrane-matrix-metalloprotease-2), beta-Nerve Growth Factor (beta-NGF), N-regulin-1 beta1/NRG-1, Tumor Necrosis Factor alpha (TNF-α), Interferon gamma-induced protein 10 (CXCL10), Interferon gamma (IFN-γ), IL-2, IL-8/CXCL8, IL-17/IL-17A, CCL-2/MCP-1 and Vascular Endothelial Growth Factor (VEGF) were analyzed. The quantification was carried out with a Bio-Plex array reader (Bio-Plex 200 System) and Bio-Plex Manager (Version 6.1 Bio-Rad Laboratories, Hercules, CA, USA) software. Student’s t or non-parametric Wilcoxon rank-sum tests were used for continuous variables to analyze the differences between groups. A p-value ≤ 0.05 was considered statistically significant. Furthermore, the receiver operating characteristic (ROC) method was used in order to find possible optimal cut-offs of the biomarkers capable of splitting patients into groups with different outcomes probabilities. Statistical analyses were conducted independently by means of SPSS® (v21.0) and MedCalc® (v10.0.1) or ‘R’ [88]. Data standardization (scaling), followed by two-way hierarchical clustering (Euclidean distance and Ward’s method was used if not specified elsewhere), were performed by means of JMP v11 (SAS Institute, Cary, NC) or ‘R’ [88] and RStudio [89]. Principal component analysis (PCA) as well as most data represented throughout the manuscript was independently reproduced by means of ‘R’ using the following packages: base, methods, utils, stats, graphics, grDevices, tcltk, openxlsx, tidyverse [90], data.table, RColorBrewer, reshape2, reshape, readxl, FactoMineR, factoextra, grid, gridExtra, circlize, cluster, dendextend and ComplexHeatmap [91]. Messenger RNA results from Taylor’s tissue dataset (NCBI GEO accession code GSE21032) have been accessed through the Prostate Cancer Genomics Data Portal (http://cbioportal/) and combined with reported clinical data. Wilcoxon/Kruskal Wallis was used to analyze the differences between groups. GraphPad Prism v4 and JMP v11 (SAS Institute, Cary, NC) were used to perform statistical analyses. All cell lines were obtained by ATCC. All cells were used as precocious (six passages) frozen stocks after arrival. They are routinely tested for Mycoplasma contamination (“PCR mycoplasma test kit”, product no. A3744, PanReac AppliChem) before EV preparations. H1299, HT1975, HT29 cells were cultivated as recommended protocols. SW480 line was maintained in RPMI and 10% of Fetal Bovine Serum (FBS). A431 and 293T cells were cultivated in DMEM (Dulbecco’s Modified Eagle’s Medium) with 10% of FBS, Glutamine (Gln) and Penicillin–Streptomycin (P/S) at standard doses. 293T cells were stable transduced with TWEEN vector [92] empty (Control) or with PD-L1 gene. Sequence-verified cDNA encoding for human PD-L1 was purchased from Dharmacon, cut with XbaI and EcoRV and inserted into TWEEN vector. It was kindly provided by Dr.Valeria Coppola. (PD-L1 Human-MGC Human CD274 Sequence-Verified cDNA-Clone ID: 30915301-Catalog Number: MHS6278-202856825) (Lentiviral manipulation authorized by Ministry of Health rules. RM/IC/Op2/17/002.notifica I.5.i.s/2017/15 - Biotecnologie. D.L.vo 206/2001). PC20 cancer activated fibroblast were obtained by tumor primary tissue cultures as by previously published [93]. Primary PCa cultures were derived from freshly-explanted tissue specimens (PCa-derived ex vivo model) following immortalization and phenotypic characterization. Clinical data and outcome of patients were collected for 15 years [66]. Briefly, poor prognosis group of donor patients with clinically localized PCa was defined by the presence of biochemical/local recurrence, metastasis, or disease-specific mortality, while the good prognosis group was defined by complete remission after surgery alone. Prognostic signature “Bad versus Good Prognosis profiling” was obtained in PCa cells by Affymetrix array (Human U133A Gene ChIP platform) using PCa cells derived from patients with different progression of disease (recurrent versus non-recurrent disease). Regulated biological processes were identified by the GOAL Web-based application and Gene Ontology (GO) terms with p < 0.01 considered differentially regulated (false discovery rate = 0.013). Affymetrix Gene Chip scanning was analyzed by customized R language-based script [88] using Bioconductor (http://www.bioconductor.org) for quality-control analysis, data normalization, hierarchical clustering, and identification of differentially expressed transcripts. Biological processes and molecular functions involved were identified by the GOAL Web-based application and the Unigene Build 154 according to the Gene Ontology (GO; http://www.geneontology.org) Consortium classification. Genes reported with p < 0.01 were considered differentially regulated (false discovery rate = 0.013 [65, 66]).Statistical analysis and data representation
Protein analyses in EV samples
Analysis of publicly available datasets
Cell lines, PCa-derived cells and gene expression profiling (Affymetrix)
Cell cultures
PCa-derived cells
Affymetrix data analysis
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clin. 2018;68:394–424.
De Angelis R, Sant M, Coleman MP, Francisci S, Baili P, Pierannunzio D, et al. Cancer survival in Europe 1999-2007 by country and age: results of EUROCARE–5-a population-based study. Lancet Oncol. 2014;15:23–34.
Walters S, Maringe C, Coleman MP, Peake MD, Butler J, Young N, et al. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: a population-based study, 2004–2007. Thorax 2013;68:551–64.
Neppl-Huber C, Zappa M, Coebergh JW, Rapiti E, Rachtan J, Holleczek B, et al. Changes in incidence, survival and mortality of prostate cancer in Europe and the United States in the PSA era: additional diagnoses and avoided deaths. Ann Oncol. 2012;23:1325–34.
O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol. 2020;21:585–606.
Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer—implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15:617–38.
Li SP, Lin ZX, Jiang XY, Yu XY. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacologica Sin. 2018;39:542–51.
Logozzi M, De Milito A, Lugini L, Borghi M, Calabrò L, Spada M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PloS ONE. 2009;4:e5219.
Sharma P, Diergaarde B, Ferrone S, Kirkwood JM, Whiteside TL. Melanoma cell-derived exosomes in plasma of melanoma patients suppress functions of immune effector cells. Sci. Rep. 2020;10:92.
Whiteside TL. The potential of tumor-derived exosomes for noninvasive cancer monitoring: an update. Expert Rev. Mol. Diagn. 2018;18:1029–40.
Alfonsi R, Grassi L, Signore M, Bonci D. The double face of exosome-carried MicroRNAs in cancer immunomodulation. Int J Mol Sci. 2018;19:1183.
Liu Y, Cao X. Organotropic metastasis: role of tumor exosomes. Cell Res. 2016;26:149–50.
Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, et al. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell 2019;177:414–27. e13.
Rajagopal C, Harikumar KB. The origin and functions of exosomes in cancer. Front Oncol. 2018;8:66.
Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, et al. Cells release prions in association with exosomes. Proc Natl Acad Sci USA. 2004;101:9683–8.
Asare-Werehene M, Nakka K, Reunov A, Chiu CT, Lee WT, Abedini MR, et al. The exosome-mediated autocrine and paracrine actions of plasma gelsolin in ovarian cancer chemoresistance. Oncogene 2020;39:1600–16.
Meldolesi J. Exosomes and ectosomes in intercellular communication. Curr Biol: CB. 2018;28:R435–r44.
Tang X, Chang C, Guo J, Lincoln V, Liang C, Chen M, et al. Tumour-secreted Hsp90alpha on external surface of exosomes mediates tumour—stromal cell communication via autocrine and paracrine mechanisms. Sci. Rep. 2019;9:15108.
Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–26.
Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015;527:329–35.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, **ao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22.
Hussain M, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Survival with olaparib in metastatic castration-resistant prostate cancer. N Engl J Med. 2020;383:2345–57.
Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, et al. Cumulative association of five genetic variants with prostate cancer. N. Engl. J. Med. 2008;358:910–9.
Lorenc T, Klimczyk K, Michalczewska I, Slomka M, Kubiak-Tomaszewska G, Olejarz W. Exosomes in prostate cancer diagnosis, prognosis and therapy. Int J Mol Sci. 2020;21:2118.
Flores-Morales A, Iglesias-Gato D. Quantitative mass spectrometry-based proteomic profiling for precision medicine in prostate cancer. Front Oncol. 2017;7:267.
Drake JM, Paull EO, Graham NA, Lee JK, Smith BA, Titz B, et al. Phosphoproteome integration reveals patient-specific networks in prostate. Cancer Cell. 2016;166:1041–54.
Malik A, Srinivasan S, Batra J. A new era of prostate cancer precision medicine. Front Oncol. 2019;9:1263.
Cannistraci A, Federici G, Addario A, Di Pace AL, Grassi L, Muto G, et al. C-Met/miR-130b axis as novel mechanism and biomarker for castration resistance state acquisition. Oncogene 2017;36:3718–28.
Vinik Y, Ortega FG, Mills GB, Lu Y, Jurkowicz M, Halperin S, et al. Proteomic analysis of circulating extracellular vesicles identifies potential markers of breast cancer progression, recurrence, and response. Sci Adv. 2020;6:eaba5714.
Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750.
Jiang L, Shen Y, Guo D, Yang D, Liu J, Fei X, et al. EpCAM-dependent extracellular vesicles from intestinal epithelial cells maintain intestinal tract immune balance. Nat Commun. 2016;7:13045.
Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009;69:5627–9.
De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharm Sci. 2019;40:172–86.
Hase T, Sato M, Yoshida K, Girard L, Takeyama Y, Horio M, et al. Pivotal role of epithelial cell adhesion molecule in the survival of lung cancer cells. Cancer Sci. 2011;102:1493–500.
Manzotti G, Torricelli F, Benedetta D, Lococo F, Sancisi V, Rossi G, et al. An epithelial-to-mesenchymal transcriptional switch triggers evolution of pulmonary sarcomatoid carcinoma (PSC) and identifies dasatinib as new therapeutic option. Clin Cancer Res. 2019;25:2348–60.
Weiswald LB, Guinebretière JM, Richon S, Bellet D, Saubaméa B, Dangles-Marie V. In situ protein expression in tumour spheres: development of an immunostaining protocol for confocal microscopy. BMC Cancer. 2010;10:106.
Fattore L, Ruggiero CF, Liguoro D, Mancini R, Ciliberto G. Single cell analysis to dissect molecular heterogeneity and disease evolution in metastatic melanoma. Cell Death Dis. 2019;10:827.
Haigler H, Ash JF, Singer SJ, Cohen S. Visualization by fluorescence of the binding and internalization of epidermal growth factor in human carcinoma cells A-431. Proc Natl Acad Sci USA. 1978;75:3317–21.
Kaplan M, Narasimhan S, de Heus C, Mance D, van Doorn S, Houben K, et al. EGFR dynamics change during activation in native membranes as revealed by NMR. Cell 2016;167:1241–51. e11.
Kriegs M, Clauditz TS, Hoffer K, Bartels J, Buhs S, Gerull H, et al. Analyzing expression and phosphorylation of the EGF receptor in HNSCC. Sci. Rep. 2019;9:13564.
Horita H, Law A, Hong S, Middleton K. Identifying regulatory posttranslational modifications of PD-L1: a focus on monoubiquitinaton. Neoplasia (New York, NY). 2017;19:346–53.
Teramoto K, Igarashi T, Kataoka Y, Ishida M, Hanaoka J, Sumimoto H, et al. Clinical significance of PD-L1-positive cancer-associated fibroblasts in pN0M0 non-small cell lung cancer. Lung Cancer (Amst., Neth.). 2019;137:56–63.
Ready N, Hellmann MD, Awad MM, Otterson GA, Gutierrez M, Gainor JF, et al. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol. 2019;37:992–1000.
Linxweiler J, Junker K. Extracellular vesicles in urological malignancies: an update. Nat Rev Urol. 2020;17:11–27.
Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016;113:E968–77.
Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977.
Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20:332–43.
Li C, Li C, Zhi C, Liang W, Wang X, Chen X, et al. Clinical significance of PD-L1 expression in serum-derived exosomes in NSCLC patients. J Transl Med. 2019;17:355.
Tomlins SA, Day JR, Lonigro RJ, Hovelson DH, Siddiqui J, Kunju LP, et al. Urine TMPRSS2:ERG plus PCA3 for individualized prostate cancer risk assessment. Eur. Urol. 2016;70:45–53.
Gevensleben H, Dietrich D, Golletz C, Steiner S, Jung M, Thiesler T, et al. The immune checkpoint regulator PD-L1 is highly expressed in aggressive primary prostate cancer. Clin. Cancer Res. 2016;22:1969–77.
Li F, Aljahdali I, Ling X. Cancer therapeutics using survivin BIRC5 as a target: what can we do after over two decades of study? J Exp Clin Cancer Res. 2019;38:368.
Culig Z, Puhr M. Interleukin-6 and prostate cancer: current developments and unsolved questions. Mol. Cell Endocrinol. 2018;462:25–30.
Le T, Gerber DE. Newer-generation EGFR inhibitors in lung cancer: how are they best used? Cancers. 2019;11:366.
Vaz J, Ansari D, Sasor A, Andersson R. SPARC: a potential prognostic and therapeutic target in pancreatic cancer. Pancreas 2015;44:1024–35.
Michalaki V, Syrigos K, Charles P, Waxman J. Serum levels of IL-6 and TNF-alpha correlate with clinicopathological features and patient survival in patients with prostate cancer. Br J Cancer. 2004;90:2312–6.
Smolarz M, Pietrowska M, Matysiak N, Mielańczyk Ł, Widłak P. Proteome profiling of exosomes purified from a small amount of human serum: the problem of co-purified serum components. Proteomes. 2019;7:18.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–w102.
Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–57.
Alonzo DG, Mure AL, Soloway MS. Prostate cancer and the increasing role of active surveillance. Postgrad Med. 2013;125:109–16.
Klotz L, Emberton M. Management of low risk prostate cancer-active surveillance and focal therapy. Nat Rev Clin Oncol. 2014;11:324–34.
Haider M, Zhang X, Coleman I, Ericson N, True LD, Lam HM, et al. Epithelial mesenchymal-like transition occurs in a subset of cells in castration resistant prostate cancer bone metastases. Clin. Exp. Metastasis. 2016;33:239–48.
Cai C, Wang H, He HH, Chen S, He L, Ma F, et al. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. J Clin Investig. 2013;123:1109–22.
Ankerst DP, Goros M, Tomlins SA, Patil D, Feng Z, Wei JT, et al. Incorporation of urinary prostate cancer antigen 3 and TMPRSS2:ERG into prostate cancer prevention trial risk calculator. Eur Urol Focus. 2019;5:54–61.
Singhal U, Wang Y, Henderson J, Niknafs YS, Qiao Y, Gursky A, et al. Multigene profiling of CTCs in mCRPC identifies a clinically relevant prognostic signature. Mol Cancer Res: MCR. 2018;16:643–54.
Nanni S, Aiello A, Re A, Guffanti A, Benvenuti V, Colussi C, et al. Estrogen-dependent dynamic profile of eNOS-DNA associations in prostate cancer. PloS ONE. 2013;8:e62522.
Nanni S, Priolo C, Grasselli A, D’Eletto M, Merola R, Moretti F, et al. Epithelial-restricted gene profile of primary cultures from human prostate tumors: a molecular approach to predict clinical behavior of prostate cancer. Mol Cancer Res: MCR. 2006;4:79–92.
LeBleu VS, Kalluri R. Exosomes as a multicomponent biomarker platform in cancer. Trends Cancer 2020;6:767–74.
Wang L, Skotland T, Berge V, Sandvig K, Llorente A. Exosomal proteins as prostate cancer biomarkers in urine: from mass spectrometry discovery to immunoassay-based validation. Eur. J. Pharm. Sci. 2017;98:80–5.
Ploussard G, Nicolaiew N, Marchand C, Terry S, Vacherot F, Vordos D, et al. Prospective evaluation of an extended 21-core biopsy scheme as initial prostate cancer diagnostic strategy. Eur. Urol. 2014;65:154–61.
Adler HL, McCurdy MA, Kattan MW, Timme TL, Scardino PT, Thompson TC. Elevated levels of circulating interleukin-6 and transforming growth factor-beta1 in patients with metastatic prostatic carcinoma. J. Urol. 1999;161:182–7.
Pidgeon GP, Tang K, Cai YL, Piasentin E, Honn KV. Overexpression of platelet-type 12-lipoxygenase promotes tumor cell survival by enhancing alpha(v)beta(3) and alpha(v)beta(5) integrin expression. Cancer Res. 2003;63:4258–67.
Zhang LY, Guo Q, Guan GF, Cheng W, Cheng P, Wu AH. Integrin Beta 5 is a prognostic biomarker and potential therapeutic target in glioblastoma. Front Oncol. 2019;9:904.
Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 2002;108:837–47.
Aicher LD, Campbell JS, Yeung RS. Tuberin phosphorylation regulates its interaction with hamartin. Two proteins involved in tuberous sclerosis. J Biol Chem. 2001;276:21017–21.
Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14.
Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–18.
Morrison CD, Parvani JG, Schiemann WP. The relevance of the TGF-β Paradox to EMT-MET programs. Cancer Lett. 2013;341:30–40.
Lei JH, Liu LR, Wei Q, Yan SB, Song TR, Lin FS, et al. Systematic review and meta-analysis of the survival outcomes of first-line treatment options in high-risk prostate cancer. Sci Rep. 2015;5:7713.
Kang JH, Jung MY, Choudhury M, Leof EB. Transforming growth factor beta induces fibroblasts to express and release the immunomodulatory protein PD-L1 into extracellular vesicles. FASEB J. 2020;34:2213–26.
Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–72.
Reyna MA, Haan D, Paczkowska M, Verbeke LPC, Vazquez M, Kahraman A, et al. Pathway and network analysis of more than 2500 whole cancer genomes. Nat. Commun. 2020;11:729.
Klotz L. The future of active surveillance. Transl Androl Urol. 2018;7:256–9.
Klotz L. Active surveillance in intermediate-risk prostate cancer. BJU Int. 2020;125:346–54.
Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell 2019;179:1033–55.
Matteoni S, Abbruzzese C, Matarrese P, De Luca G, Mileo AM, Miccadei S, et al. The kinase inhibitor SI113 induces autophagy and synergizes with quinacrine in hindering the growth of human glioblastoma multiforme cells. J Exp Clin Cancer Res. 2019;38:202.
Shively S, Miller WR. The use of HMDS (hexamethyldisilazane) to replace Critical Point Drying (CPD) in the preparation of tardigrade for SEM (Scanning Electron Microscope) imaging. Trans Kans Acad Sci. (1903). 2009;112:198–200.
Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3 22.
R: A Language and Environment for Statistical Computing [Internet]. R Core Team. 2018. Available from: http://www.R-project.org/.
RStudio: Integrated Development Environment for R [Internet]. RStudio Team. 2020. Available from: http://www.rstudio.com/.
Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4:1686.
Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinform (Oxf, Engl). 2016;32:2847–9.
Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–7.
Musumeci M, Coppola V, Addario A, Patrizii M, Maugeri-Sacca M, Memeo L, et al. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene 2011;30:4231–42.
Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput Graph. 2014;20:1983–92.
Acknowledgements
The authors thank Giuseppe Loreto, Paola Di Matteo, Roberto Ricci, Laura De Salvo, Marco Varmi and Mustapha Haoui for their technical support. Claudio Tabolacci was supported by Fondazione Umberto Veronesi, which is gratefully acknowledged.
Funding
The present study was supported by: National Ministry of Health, Under-forty researcher (GR:2011-02351557) to D.B., S.N., and A.G.; Oncotecnologico program 15ONC/5 to D.B.; Transcan-2-JTC 2016-Prolipsy and PON “BiLiGeCT” ARS01_00492 to D.B.
Author information
Authors and Affiliations
Contributions
M.S., R.A., G.F., A.A.D. performed RPPA and Western blotting analysis and E.V. sample preparation; M.S. and G.F. performed database data analysis; S.N., A.A.I. executed primary culture experiment analysis; LBE achieved TEM, SEM, and IEM analyses; M.D. made TEM experiments; I.S. and M.S. performed statistical analysis; A.L.D.P. and L.B. realized E.V. sample preparation, FACS analysis, ELEXO optimization, and experiments; G.M., A.G., D.C., G.S., M.C., L.C., M.G., R.P. enrolled patient cohorts, collected samples, and clinical information databases, and promoted the trial clinical design; S.S. performed anatomic pathology evaluation of the enrolled cohorts; S.R., C.T. performed Luminex experiments; T.M. performed English language revision; M.S. compiled the figures; D.B., R.D.M. coordinated and proposed the trial; D.B., M.S., R.D.M. created the experimental design and clinical sets; D.B., M.S. and R.D.M. wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All patients’ samples were collected and handled in the study following the ethical internationally recognized guidelines and the project was approved by the Ethics Committees. Prospective training-like and advanced cancer pivotal cohorts were collected at San Giovanni Bosco and Humanitas, Turin hospitals and Regina Elena National Cancer Institute under the approval of Istituto Superiore di Sanità (CE-ISS 09-250, 2009; Prot. CE-ISS-PRE 17/18 of 11/01/2018; CE-IRE-350-13). Retrospective prostate and colon adenocarcinoma cases were collected at Regina Elena National Cancer Institute under the Italy-USA program and subjected to approval by Istituto Superiore di Sanità (Prot-PRE 202/06-ref-CE-ISS 06/140). NSCL-cancer were collected under Ethics Committees approval (Prot-ISS-PRE-416/17).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Melino.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Signore, M., Alfonsi, R., Federici, G. et al. Diagnostic and prognostic potential of the proteomic profiling of serum-derived extracellular vesicles in prostate cancer. Cell Death Dis 12, 636 (2021). https://doi.org/10.1038/s41419-021-03909-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-021-03909-z
- Springer Nature Limited
This article is cited by
-
Flow-dependent shear stress affects the biological properties of pericyte-like cells isolated from human dental pulp
Stem Cell Research & Therapy (2023)
-
Extracellular vesicle-based liquid biopsy biomarkers and their application in precision immuno-oncology
Biomarker Research (2023)
-
Chlorpromazine affects glioblastoma bioenergetics by interfering with pyruvate kinase M2
Cell Death & Disease (2023)
-
Proteomics technologies for cancer liquid biopsies
Molecular Cancer (2022)
-
Serine and one-carbon metabolisms bring new therapeutic venues in prostate cancer
Discover Oncology (2021)