Abstract
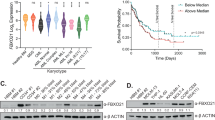
The ubiquitin–proteasome system (UPS) participates in both physiological and pathological processes through the posttranslational regulation of intracellular signal transduction pathways. F-box and WD-40 domain protein 11 (Fbxw11) is a component of the SCF (Skp1–Cul1–F-box) E3 ubiquitin ligase complex. Fbxw11 regulates various signal transduction pathways, and it may have pathological roles in tumorigenesis. However, the role of Fbxw11 in the development of leukemia and the underlying mechanisms remain largely unknown. In this study, Fbxw11 expression was aberrantly upregulated in patients with lymphocytic leukemia. Its expression was dramatically decreased in patients who achieved complete remission (CR) after chemotherapy. The high level of Fbxw11 expression in L1210 lymphocytic leukemia cells stimulated cell proliferation in vitro and tumor formation in vivo. The effects were mediated by the stimulation of cell cycle progression rather than the induction of apoptosis. Furthermore, a bioinformatics analysis suggested concomitant activation of the NF-κB and β-catenin/TCF signaling pathways, which were confirmed by reporter gene assays. Moreover, blocking experiments suggested the involvement of both pathways in the growth-promoting effects of Fbxw11. Our results reveal the role of Fbxw11 in lymphocytic leukemia cells and imply that Fbxw11 may serve as a potential molecular target for the treatment of lymphocytic leukemia.
Similar content being viewed by others
Introduction
Hematopoiesis is strictly regulated by complicated intercellular communication from the hematopoietic microenvironment through sophisticated signal transduction networks. Dysregulation of signal transduction will disrupt the balance of normal hematopoiesis and cause various blood diseases. Leukemia is regarded as a clonal disease1,2, and many intrinsic and extrinsic factors have been verified to play parts in the initiation and development of leukemia3,4. During leukemogenesis, leukemia cells outcompete their normal counterparts and become dominant due to their high capacity for self-renewal and low capacity for apoptosis5. Diverse intrinsic abnormalities, which endow leukemia cells with those characteristics, have been elucidated at different levels, including mRNA transcriptional control, protein translation, and posttranslational modifications6,7,8,9,10.
The ubiquitin–proteasome system (UPS), which is the main pathway for the degradation of short-period proteins in cells, is involved in the posttranslational regulation of numerous intracellular signal transduction pathways11. The Skp1/cullin/F-box (SCF) complex is an important E3 ubiquitin ligase. F-box family proteins, which are further divided into Fbxw, Fbxl, and Fbxo subfamilies based on protein structure, determine the specificity of substrate degradation by identifying and binding to different target proteins12. Abnormal expression or dysfunction of several F-box proteins results in aberrant ubiquitination, inducing the development and progression of malignancies, including hematopoietic malignancies. Some F-box family members contribute to tumorigenesis and tumor development13,14,15. Fbxw7 controls leukemia-initiating cells in chronic myelogenous leukemia and chronic myeloid leukemia (CML) by regulating c-Myc ubiquitination16,17,18. Fbxo11 loss or mutation induces impairments in BCL6 degradation, and therefore BCL6 accumulation contributes to pathogenesis of diffuse large B-cell lymphomas19. In addition, the E3 ligase family members Fbxl2, Fbxl10, and SKP2 participate in the proliferation of leukemia cells by regulating the ubiquitination pathway20,21,22. To date, the effects of other members in the F-box family on the development of hematopoietic malignancies have not been established.
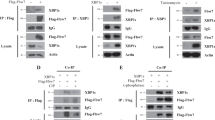
F-box and WD repeat domain containing 11 (Fbxw11), also known as HOS or β-TrCP2, belongs to the Fbxw subfamily of the F-box protein family23. Fbxw11 is crucial for embryonic development, and the most obvious defect in Fbxw11−/− mice is embryonic mortality24. Fbxw11 plays pivotal roles in various signaling pathways by regulating the ubiquitination of phosphorylated substrates. The SCFFbxw11 complex regulates many important biological processes, including the cell cycle, differentiation, development, and metabolism, by targeting a broad range of substrates, including IκB, β-catenin, ATF4, Emi1, etc.25,Full size image
Fbxw11 promoted the expression of some proliferation-associated genes
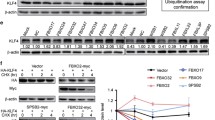
Based on the results of the GO analysis of biological processes, the category associated with proliferation was selected for further analysis, and the selected genes are listed in Fig. 5a. Thirteen of these genes were reported to stimulate proliferation, and 9 were reported to suppress proliferation in previous studies. Plac8, Nanog, Emp2, Fasl, Tnfsf18, Arg1, Gabbr1, and Plxnb1 were selected and verified by real-time PCR. With the exception of Arg1 in L1210-Fbxw11a cells, the expression of proliferation-promoting genes was upregulated while the expression of proliferation-suppressing genes was downregulated in L1210-Fbxw11a/c/d cells (Fig. 5b). These results were consistent with the gene array data. Cell cycle checkpoint genes were also analyzed to further examine the effects of Fbxw11 on the cell cycle. Most of these genes displayed low fold changes in the gene array data (Fig. 5c). However, levels of the Cyclin D1 protein, which is regulated by multiple transcription factors, including NF-κB and β-catenin, were significantly increased in L1210 cells expressing high levels of Fbxw11 transcript variants, according to the Western blot analysis (Fig. 5d). We designed a cyclin D1 shRNA to reverse of the effect of Fbxw11 on proliferation and validate the role of Cyclin D1 on the Fbxw11-induced increase in cell proliferation (Fig. 5e). As expected, knockdown of cyclin D1 decreased the proliferation of L1210 cells (Fig. 5f).
a Proliferation-associated genes, including 13 proliferation-promoting genes and 9 proliferation-suppressing genes, were selected. The data were standardized to L1210-control cells. b The expression of these genes was validated by real-time PCR. c Genes regulating the cell cycle were also selected for clustering and normalized to L1210-control cells. d Western blots were performed to detect the expression of eight integral proteins regulating the cell cycle. GAPDH was used as control. e Real-time PCR analysis showed that transfection of cells with Cyclin D1-specific shRNAs significantly reduced its mRNA levels in L1210 cells. f Cyclin D1 silencing decreased the proliferation of L1210 cells over-expressing Fbxw11c. Data are presented as means ± SEM (n = 3; 3 independent experiments). *p < 0.05, **p < 0.01, ***p < 0.001
Fbxw11 might promote cell proliferation by activating both the NF-κB and β-catenin/TCF signaling pathways
As Cyclin D1 is downstream of both the NF-κB and β-catenin/TCF signaling pathways, we must determine which pathway is activated during the Fbxw11-mediated increase in proliferation. Hence, DEGs downstream of either NF-κB or β-catenin/TCF signaling pathway are plotted in Fig. 6a, b, respectively. The genes upregulated by Fbxw11 were summarized. Among these genes, upregulation of seven NF-κB-regulated genes in L1210-Fbxw11a/c/d cells was verified by real-time PCR, although slight differences in magnitude were observed among the three groups (Fig. 6c). Eight β-catenin/TCF-regulated genes were also verified by real-time PCR. The expression of Emp2, Lgr5, and Wisp1 was upregulated in L1210-Fbxw11a/c/d cells. Dll1, Ecel1, Nanog, Cldn1, and Trx3 were upregulated to various extents in L1210 cells expressing high levels of Fbxw11 transcript variants (Fig. 6d). A dual luciferase reporter system was also used. Activation of both the NF-κB and β-catenin/TCF signaling pathways was detected in 293T cells expressing high levels of Fbxw11 transcript variants (Fig. 6e, f). ICG-001 and JSH-23, inhibitors of the NF-κB and β-catenin/TCF pathways, were used to further confirm that both the NF-κB and β-catenin/TCF signaling pathways mediate the pro-proliferation effects on L1210-Fbxw11a/c/d cells. The administration of high doses of both inhibitors (5 μM for ICG-001 and 16 μM for JSH-23) completely diminished the pro-proliferation effects of Fbxw11 on L1210 cells. Furthermore, the administration of a combination of low doses of inhibitors (2 μM for ICG-001 and 8 μM for JSH-23) completely diminished the pro-proliferation effects of Fbxw11, although treatment with a low dose of either inhibitor had little effect (Fig. 6g). Based on the results of the apoptosis analysis, all dosages used in the experiments displayed little toxic effects (Figure S4). The above data suggested that Fbxw11 promoted the proliferation of L1210 cells by concomitantly activating the NF-κB and β-catenin/TCF signaling pathways.
a, b Genes downstream of the NF-κB and β-catenin/TCF signaling pathways were selected for clustering and normalized to L1210-control cells. c, d The expression of representative genes regulated by the NF-κB and β-catenin/TCF signaling pathways were verified by real-time PCR. e The dual luciferase method was used to analyze the activation of the NF-κB pathway in HEK293T cells expressing high levels of the Fbxw11 transcript variants. The control was set to 1. f The dual luciferase method was used to analyze the activation of the β-catenin/TCF signaling pathway in HEK293T cells expressing high levels of Fbxw11 transcript variants, and a mutant TCF transcription factor (FOP) was used as a functional control. The controls were set to 1. g The effects of NF-κB and β-catenin/TCF signaling pathway inhibitors on the proliferation of leukemia cells. Data are presented as means ± SEM (n = 3; 3 independent experiments). *Compared to control cells, *p < 0.05, **p < 0.01, ***p < 0.001; #Compared to DMSO, #p < 0.05, ###p < 0.001