Abstract
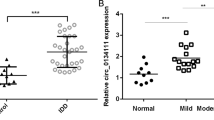
Intervertebral disc degeneration (IDD) is an important factor leading to low back pain, but the underlying mechanisms remain poorly understood. Compared with normal nucleus pulposus (NP) tissues, the expression of circ-GRB10 was downregulated in IDD. Furthermore, overexpression of circ-GRB10 inhibited NP cell apoptosis. circ-GRB10 could sequester miR-328-5p, which could potentially lead to the upregulation of target genes related to cell proliferation via the ErbB pathway. In conclusion, the present study revealed that circ-GRB10/miR-328-5p/ERBB2 signaling pathway is involved in IDD development, suggesting that circ-GRB10 might be a novel therapeutic target for IDD.
Similar content being viewed by others
Introduction
Intervertebral disc degeneration (IDD) is characterized by extracellular matrix breakdown and abnormal matrix synthesis leading to reduced hydration, loss of disc height, and decreased potential to absorb load1,2. It is considered as the predominant cause for chronic low back pain and spine-related disease, leading to economic and social burden worldwide3. It is estimated that as much as 84% of the population suffers from low back pain at some point in their lifetime, whereas 10% are chronically disabled4. Nevertheless, current strategies for IDD treatment is hampered by an incomplete understanding of its pathogenesis. Nowadays, IDD treatment is limited to symptomatic interventions, which do not adequately improve outcomes since no disease-modifying drugs are available5. Consequently, the clinical management of diseases related to IDD remains severely limited. Therefore, unveiling the pathophysiology and molecular mechanisms underlying IDD is imperative for develo** novel therapeutic approaches.
The intervertebral disc is the largest avascular structure in the body6. Studies have shown that blood vessels do not enter the nucleus pulposus (NP) tissue, and NP cells in the center of an adult disk can be up to 8 mm away from the nearest blood vessel7. Accumulating evidence indicated that a variety of cellular events are dysregulated in the progression of IDD, ranging from NP cell apoptosis to cytokine expression7,8,9,Full size image
Circ-GRB10 regulates the expression of ERBB2
Bioinformatics prediction showed that ERBB2 was the target of miR-328-5p, as predicted by three microRNA target gene databases (DIANA, miRDB, and miRTarbase) (Fig. 6a). To determine whether circ-GRB10 could regulate ERBB2 expression through miR-328-5p, we analyzed the mRNA and protein levels of ERBB2 in NP cells isolated from normal tissues. The expression of circ-GRB10 was attenuated after siRNA silencing (P < 0.001) and increased by circ-GRB10 overexpression (P < 0.001) (Supplemental Fig. 3a). The expression of miR-328-5p was upregulated after miR-328-5p mimic addition (P < 0.001) and downregulated after miR-328-5p inhibitor treatment (P < 0.01) (Supplemental Fig. 3b). Moreover, the expression of miR-328-5p was altered after circ-GRB10 silencing or overexpression (Supplemental Fig. 3c). As shown in Fig. 6b, c, the expression of ERBB2 was inhibited by miR-328-5p mimics and could be reversed by circ-GRB10 overexpression at both the mRNA and protein levels. Circ-GRB10 silencing resulted in a reduction of ERBB2 at both the mRNA and protein levels, as well as in the downregulation of miR-328-5p. Meanwhile, the expression of ERBB2 was elevated in NP cells treated with a miR-328-5p inhibitor (Fig. 6d, e). These results indicated that circ-GRB10 regulates ERBB2 through miR-328-5p.
a The Venn diagram indicates the relevant target genes mediated by miR-328-5p. ERBB2 was intersected and predicted by three different databases. b NP cells from control tissues were transfected with miR-328-5p with or without circ-GRB10 overexpress plasmid. qRT-PCR was used to detect the relative mRNA levels of ERBB2 compared with controls. c Relative protein levels of ERBB2 when transfected with miR-328-5p mimics and reversed by circ-GRB10 expression plasmid. d circ-GRB10 siRNA with or without miR-328-5p inhibitor was transfected into NP cells from control tissues and the mRNA level of ERBB2 was evaluated by qRT-PCR. e Western blot analysis of ERBB2 protein level following treatment of NP cells from control tissues with circ-GRB10 siRNA or miR-328-5p inhibitor. GAPDH was used as control. *P < 0.05, **P < 0.01, n = 3
Circ-GRB10-miR-328-5p-ERBB2 regulatory loop is critical for NP cells survival
As shown in Fig. 7a, b, the cell death rate in IDD NP tissues was significantly higher than in normal NP cells, as indicated by the TUNEL assay(P < 0.01). We next checked whether circ-GRB10 participated in NP cell apoptosis and survival. After nutrition deprivation, the constitutive expression of circ-GRB10 and ERBB2 was downregulated compared to normal cells (all P < 0.01, Fig. 8a, b). The apoptosis in NPs cells of the circ-GRB10 overexpression group was significantly suppressed compared with the mock circRNA vector group (P < 0.001); nevertheless, circ-GRB10 knockdown markedly increased the NP cell apoptosis rate (P < 0.001); circ-GRB10 could significantly stimulate NP cells survival under nutrient deprivation conditions (P < 0.01) (Fig. 8h). Moreover, the expression of apoptosis-related proteins was measured. As shown in Fig. 8k, i, the expression of cleaved caspase-3 was upregulated after circ-GRB10 silencing and by nutrient deprivation (both P < 0.001), and attenuated by circ-GRB10 overexpression (P < 0.05). Figure 8m, n shows that the expression of cleaved PARP1 was upregulated after circ-GRB10 silencing and by nutrient deprivation (both P < 0.001), and attenuated by circ-GRB10 overexpression (P < 0.01). The LC3-II/LC3-I ratio was increased after circ-GRB10 silencing and by nutrient deprivation (both P < 0.001), and decreased by circ-GRB10 overexpression (P < 0.01) (Fig. 8o, p). Moreover, Fig. 8q, r shows that the expression of p62 was upregulated after circ-GRB10 silencing and by nutrient deprivation (both P < 0.001), and attenuated by circ-GRB10 overexpression (P < 0.001).
a, b Constitutive expression levels of circ-GRB10 and ERBB2 protein in nutrient deprivation condition as well as normal ones. c Blank group. d Scrambled siRNA group. e circ-GRB10 si group. f Mock circular vector circRNA group. g circ-GRB10 overexpression (circ-GRB10 OE) group. h nutrient deprivation group. i Function recovery group, i.e., nutrient deprivation + circ-GRB10 OE group. j Apoptosis rate of each rate. k, l Expression of C-caspase3. m, n Expression of C-PARP1. o, p LC3-II/LC3-I ratios. q, r Expression of p62. (**P < 0.01, ***P < 0.001, n = 3. C-caspase3, cleaved caspase3; C-PARP1, cleaved PARP1)