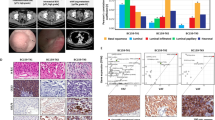
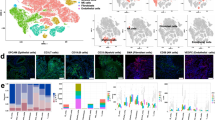
Abstract
Metastasis is the greatest clinical challenge for UTUCs, which may have distinct molecular and cellular characteristics from earlier cancers. Herein, we provide single-cell transcriptome profiles of UTUC para cancer normal tissue, primary tumor lesions, and lymphatic metastases to explore possible mechanisms associated with UTUC occurrence and metastasis. From 28,315 cells obtained from normal and tumor tissues of 3 high-grade UTUC patients, we revealed the origin of UTUC tumor cells and the homology between metastatic and primary tumor cells. Unlike the immunomicroenvironment suppression of other tumors, we found no immunosuppression in the tumor microenvironment of UTUC. Moreover, it is imperative to note that stromal cells are pivotal in the advancement of UTUC. This comprehensive single-cell exploration enhances our comprehension of the molecular and cellular dynamics of metastatic UTUCs and discloses promising diagnostic and therapeutic targets in cancer-microenvironment interactions.









Similar content being viewed by others
Data availability
The data used to support the findings of this study are available from the corresponding authors upon request.
Code availability
The code used to support the findings of this study is available upon request from the corresponding author.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. https://doi.org/10.3322/caac.21708.
Hassler MR, Bray F, Catto JWF, Grollman AP, Hartmann A, Margulis V, et al. Molecular characterization of upper tract urothelial carcinoma in the era of next-generation sequencing: a systematic review of the current literature. Eur Urol. 2020;78 :209–20. https://doi.org/10.1016/j.eururo.2020.05.039.
Crivelli JJ, Xylinas E, Kluth LA, Rieken M, Rink M, Shariat SF. Effect of smoking on outcomes of urothelial carcinoma: a systematic review of the literature. Eur Urol. 2014;65:742–54. https://doi.org/10.1016/j.eururo.2013.06.010.
Van Osch FH, Jochems SH, van Schooten FJ, Bryan RT, Zeegers MP. Significant role of lifetime cigarette smoking in worsening bladder cancer and upper tract urothelial carcinoma prognosis: a meta-analysis. J Urol. 2016;195:872–9. https://doi.org/10.1016/j.juro.2015.10.139.
Rouprêt M, Babjuk M, Burger M, Capoun O, Cohen D, Compérat EM, et al. European Association of Urology Guidelines on upper urinary tract urothelial carcinoma: 2020 update. Eur Urol. 2021;79:62–79. https://doi.org/10.1016/j.eururo.2020.05.042.
Laplane L, Duluc D, Bikfalvi A, Larmonier N, Pradeu T. Beyond the tumour microenvironment [published correction appears in Int J Cancer. 2021 Mar 15;148(6):E5]. Int J Cancer. 2019;145:2611–8. https://doi.org/10.1002/ijc.32343.
Huang C, Bruggeman LA, Hydo LM, Miller RT. Shear stress induces cell apoptosis via a c-Src-phospholipase D-mTOR signaling pathway in cultured podocytes. Exp Cell Res. 2012;318:1075–85. https://doi.org/10.1016/j.yexcr.2012.03.011.
Ahn S, Park H. XIAP is essential for shear stress-enhanced Tyr-576 phosphorylation of FAK. Biochem Biophys Res Commun. 2010;399:256–61. https://doi.org/10.1016/j.bbrc.2010.07.064.
McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–28. https://doi.org/10.1016/j.cell.2017.01.018.
McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. https://doi.org/10.1016/j.ccell.2014.12.001.
Blanpain C. Tracing the cellular origin of cancer. Nat Cell Biol. 2013;15:126–34. https://doi.org/10.1038/ncb2657.
Lourenco AR, Ban Y, Crowley MJ, Lee SB, Ramchandani D, Du W, et al. Differential contributions of pre- and post-EMT tumor cells in breast cancer metastasis. Cancer Res. 2020;80:163–9. https://doi.org/10.1158/0008-5472.CAN-19-1427.
Davis RT, Blake K, Ma D, Gabra MBI, Hernandez GA, Phung AT, et al. Transcriptional diversity and bioenergetic shift in human breast cancer metastasis revealed by single-cell RNA sequencing. Nat Cell Biol. 2020;22:310–20. https://doi.org/10.1038/s41556-020-0477-0.
Müller M, Bird TG, Nault JC. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J Hepatol. 2020;72:990–1002. https://doi.org/10.1016/j.jhep.2020.01.019.
Rebouissou S, Nault JC. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J Hepatol. 2020;72:215–29. https://doi.org/10.1016/j.jhep.2019.08.017.
Baslan T, Hicks J. Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat Rev Cancer. 2017;17:557–69. https://doi.org/10.1038/nrc.2017.58.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401. https://doi.org/10.1126/science.1254257.
Albini A, Sporn MB. The tumour microenvironment as a target for chemoprevention. Nat Rev Cancer. 2007;7:139–47. https://doi.org/10.1038/nrc2067.
Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–37. https://doi.org/10.1038/nm.3394.
Hosein AN, Huang H, Wang Z, Parmar K, Du W, Huang J, et al. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight. 2019;5:e129212. https://doi.org/10.1172/jci.insight.129212.
Wirtz D, Konstantopoulos K, Searson PC. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer. 2011;11:512–22. https://doi.org/10.1038/nrc3080.
Turajlic S, Swanton C. Metastasis as an evolutionary process. Science. 2016;352:169–75. https://doi.org/10.1126/science.aaf2784.
Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. https://doi.org/10.1038/ng1060.
Chen YC, Sahoo S, Brien R, Jung S, Humphries B, Lee W, et al. Single-cell RNA-sequencing of migratory breast cancer cells: discovering genes associated with cancer metastasis. Analyst. 2019;144:7296–309. https://doi.org/10.1039/c9an01358j.
Kim KT, Lee HW, Lee HO, Song HJ, Jeong da E, Shin S, et al. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol. 2016;17:80. https://doi.org/10.1186/s13059-016-0945-9.
Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–49.e21. https://doi.org/10.1016/j.cell.2019.06.024.
Liang Y, Tan Y, Guan B, Guo B, **a M, Li J, et al. Single-cell atlases link macrophages and CD8+ T-cell subpopulations to disease progression and immunotherapy response in urothelial carcinoma. Theranostics. 2022;12:7745–59. https://doi.org/10.7150/thno.77281.
Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–98. https://doi.org/10.1038/nrc.2016.73.
Ajani JA, Wang X, Song S, Suzuki A, Taketa T, Sudo K, et al. ALDH-1 expression levels predict response or resistance to preoperative chemoradiation in resectable esophageal cancer patients. Mol Oncol. 2014;8:142–9. https://doi.org/10.1016/j.molonc.2013.10.007.
Patterson LH, Murray GI. Tumour cytochrome P450 and drug activation. Curr Pharm Des. 2002;8:1335–47. https://doi.org/10.2174/1381612023394502.
Casazza A, Laoui D, Wenes M, Rizzolio S, Bassani N, Mambretti M, et al. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell. 2013;24:695–709. https://doi.org/10.1016/j.ccr.2013.11.007.
Tanaka K, Arao T, Maegawa M, Matsumoto K, Kaneda H, Kudo K, et al. SRPX2 is overexpressed in gastric cancer and promotes cellular migration and adhesion. Int J Cancer. 2009;124:1072–80. https://doi.org/10.1002/ijc.24065.
Liu KL, Wu J, Zhou Y, Fan JH. Increased Sushi repeat-containing protein X-linked 2 is associated with progression of colorectal cancer. Med Oncol. 2015;32:99. https://doi.org/10.1007/s12032-015-0548-4.
Gao Z, Zhang J, Bi M, Han X, Han Z, Wang H, et al. SRPX2 promotes cell migration and invasion via FAK dependent pathway in pancreatic cancer. Int J Clin Exp Pathol. 2015;8:4791–8.
Tang H, Zhao J, Zhang L, Zhao J, Zhuang Y, Liang P. SRPX2 enhances the epithelial-mesenchymal transition and temozolomide resistance in glioblastoma cells. Cell Mol Neurobiol. 2016;36:1067–76. https://doi.org/10.1007/s10571-015-0300-9.
Horie M, Saito A, Yamaguchi Y, Ohshima M, Nagase T. Three-dimensional Co-culture model for tumor-stromal interaction. J Vis Exp. 2015;96:52469. https://doi.org/10.3791/52469.
Gonzalez-Zubeldia I, Dotor J, Redrado M, Bleau AM, Manrique I, de Aberasturi AL, et al. Co-migration of colon cancer cells and CAFs induced by TGFβ1 enhances liver metastasis. Cell Tissue Res. 2015;359:829–39. https://doi.org/10.1007/s00441-014-2075-6.
Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–30. https://doi.org/10.1038/nature21349.
Smeland S, Bielack SS, Whelan J, Bernstein M, Hogendoorn P, Krailo MD, et al. Survival and prognosis with osteosarcoma: outcomes in more than 2000 patients in the EURAMOS-1 (European and American Osteosarcoma Study) cohort. Eur J Cancer. 2019;109:36–50. https://doi.org/10.1016/j.ejca.2018.11.027.
Harris RJ, Cheung A, Ng JCF, Laddach R, Chenoweth AM, Crescioli S, et al. Tumor-infiltrating B lymphocyte profiling identifies IgG-biased, clonally expanded prognostic phenotypes in triple-negative breast cancer. Cancer Res. 2021;81:4290–304. https://doi.org/10.1158/0008-5472.CAN-20-3773.
Magerus A, Bercher-Brayer C, Rieux-Laucat F. The genetic landscape of the FAS pathway deficiencies. Biomed J. 2021;44:388–99. https://doi.org/10.1016/j.bj.2021.06.005.
Vonderheide RH. CD40 agonist antibodies in cancer immunotherapy. Annu Rev Med. 2020;71:47–58. https://doi.org/10.1146/annurev-med-062518-045435.
Brantley-Sieders DM, Caughron J, Hicks D, Pozzi A, Ruiz JC, Chen J. EphA2 receptor tyrosine kinase regulates endothelial cell migration and vascular assembly through phosphoinositide 3-kinase-mediated Rac1 GTPase activation. J Cell Sci. 2004;117:2037–49. https://doi.org/10.1242/jcs.01061.
Stewart GD, Bariol SV, Grigor KM, Tolley DA, McNeill SA. A comparison of the pathology of transitional cell carcinoma of the bladder and upper urinary tract. BJU Int. 2005;95:791–3. https://doi.org/10.1111/j.1464-410X.2005.05402.x.
Cha EK, Shariat SF, Kormaksson M, Novara G, Chromecki TF, Scherr DS, et al. Predicting clinical outcomes after radical nephroureterectomy for upper tract urothelial carcinoma. Eur Urol. 2012;61:818–25. https://doi.org/10.1016/j.eururo.2012.01.021.
Sobierajska K, Ciszewski WM, Sacewicz-Hofman I, Niewiarowska J. Endothelial cells in the tumor microenvironment. Adv Exp Med Biol. 2020;1234:71–86. https://doi.org/10.1007/978-3-030-37184-5_6.
Kuzet SE, Gaggioli C. Fibroblast activation in cancer: when seed fertilizes soil. Cell Tissue Res. 2016;365:607–19. https://doi.org/10.1007/s00441-016-2467-x.
Dong D, Yao Y, Song J, Sun L, Zhang G. Cancer-associated fibroblasts regulate bladder cancer invasion and metabolic phenotypes through autophagy. Dis Markers. 2021;2021:6645220 https://doi.org/10.1155/2021/6645220.
Chen Z, Zhou L, Liu L, Hou Y, **ong M, Yang Y, et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun. 2020;11:5077. https://doi.org/10.1038/s41467-020-18916-5.
Liu B, Pan S, Liu J, Kong C. Cancer-associated fibroblasts and the related Runt-related transcription factor 2 (RUNX2) promote bladder cancer progression. Gene. 2021;775:145451. https://doi.org/10.1016/j.gene.2021.145451.
Chung W, Eum HH, Lee HO, Lee KM, Lee HB, Kim KT, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081 https://doi.org/10.1038/ncomms15081.
**a W, Zhang S, Duan H, Wang C, Qian S, Shen H. The combination therapy of Everolimus and anti-PD-1 improves the antitumor effect by regulating CD8+ T cells in bladder cancer. Med Oncol. 2022;39:37. https://doi.org/10.1007/s12032-021-01624-5.
Oh DY, Kwek SS, Raju SS, Li T, McCarthy E, Chow E, et al. Intratumoral CD4+ T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell. 2020;181:1612–25.e13. https://doi.org/10.1016/j.cell.2020.05.017.
Horn T, Laus J, Seitz AK, Maurer T, Schmid SC, Wolf P, et al. The prognostic effect of tumour-infiltrating lymphocytic subpopulations in bladder cancer. World J Urol. 2016;34:181–7. https://doi.org/10.1007/s00345-015-1615-3.
Wang T, Zhou Q, Zeng H, Zhang H, Liu Z, Shao J, et al. CCR8 blockade primes anti-tumor immunity through intratumoral regulatory T cells destabilization in muscle-invasive bladder cancer. Cancer Immunol Immunother. 2020;69:1855–67. https://doi.org/10.1007/s00262-020-02583-y.
Winerdal ME, Krantz D, Hartana CA, Zirakzadeh AA, Linton L, Bergman EA, et al. Urinary bladder cancer tregs suppress MMP2 and potentially regulate invasiveness. Cancer Immunol Res. 2018;6:528–38. https://doi.org/10.1158/2326-6066.CIR-17-0466.
Goc J, Germain C, Vo-Bourgais TK, Lupo A, Klein C, Knockaert S, et al. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res. 2014;74:705–15. https://doi.org/10.1158/0008-5472.CAN-13-1342.
Nazarkina ZhK, Laktionov PP. [Preparation of dendritic cells for cancer immunotherapy]. Biomed Khim. 2015;61:30–40. https://doi.org/10.18097/pbmc20156101030.
Laoui D, Keirsse J, Morias Y, Van Overmeire E, Geeraerts X, Elkrim Y, et al. The tumour microenvironment harbours ontogenically distinct dendritic cell populations with opposing effects on tumour immunity. Nat Commun. 2016;7:13720. https://doi.org/10.1038/ncomms13720.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (Grant Nos.: 81972409 and 81672549) and the Medical Science and Technology Project of Henan Province in 2019 (Grant No.: LHGJ20190567) and 2020 (Grant No.: LHGJ20200582).
Author information
Authors and Affiliations
Contributions
Lin Ye designed the research methods. Shiyong **n, Zhenhua Zhang, and **anchao Sun performed the analysis. Liang ** and Weiyi Li analyzed the data. Shiyong **n drafted and revised the manuscript. All authors approved the final version of the submitted manuscript and agreed to be responsible for all aspects of the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
**n, S., Zhang, Y., Zhang, Z. et al. ScRNA-seq revealed the tumor microenvironment heterogeneity related to the occurrence and metastasis in upper urinary tract urothelial carcinoma. Cancer Gene Ther (2024). https://doi.org/10.1038/s41417-024-00779-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41417-024-00779-3
- Springer Nature America, Inc.