Abstract
In humans, genetic variants of DLGAP1-4 have been linked with neuropsychiatric conditions, including autism spectrum disorder (ASD). While these findings implicate the encoded postsynaptic proteins, SAPAP1-4, in the etiology of neuropsychiatric conditions, underlying neurobiological mechanisms are unknown. To assess the contribution of SAPAP4 to these disorders, we characterized SAPAP4-deficient mice. Our study reveals that the loss of SAPAP4 triggers profound behavioural abnormalities, including cognitive deficits combined with impaired vocal communication and social interaction, phenotypes reminiscent of ASD in humans. These behavioural alterations of SAPAP4-deficient mice are associated with dramatic changes in synapse morphology, function and plasticity, indicating that SAPAP4 is critical for the development of functional neuronal networks and that mutations in the corresponding human gene, DLGAP4, may cause deficits in social and cognitive functioning relevant to ASD-like neurodevelopmental disorders.
Similar content being viewed by others
Introduction
Throughout life, several dynamic processes control the morphology, number and strength of brain synapses. Alterations in these parameters are mediated through changes in the molecular composition of synapses and via chemical modification of synaptic proteins. Excitatory brain synapses are characterized by the so-called postsynaptic density (PSD), a dense array of more than a thousand different proteins, including transmembrane receptors and adhesion molecules, scaffold and adapter proteins, cytoskeletal components and signalling molecules that together form a dynamic macromolecular complex1,2,3,4. Tight control of the molecular composition of PSDs is required to establish and maintain proper functional brain circuitry underlying cognition and behaviour. The PSD core contains several modular scaffold proteins of four different families, the Dlg/PSD-95/SAP90 (here collectively referred to as Dlg), SAPAP/GKAP (referred to as SAPAP), Shank/ProSAP (referred to as Shank) and Homer family. Dlg proteins directly associate with ionotropic glutamate receptors (iGluRs) and SAPAPs link these complexes to Shank proteins. The latter build a second molecular layer within PSDs and interact with microfilaments and Homer proteins, which then associate with metabotropic glutamate receptors (mGluRs). This physical link appears to facilitate a functional crosstalk between iGluRs and mGluRs and creates a central platform that organizes the molecular composition of and the signalling within the PSD.
In humans, mutations in genes encoding synaptic proteins have been linked with neurodevelopmental disorders, suggesting that the phenotypic expression of these disorders is, at least in part, attributed to dysfunctions of synapses and neuronal networks2,5,6,7,8,9,10. Affected proteins include presynaptic components, trans-synaptic adhesion/signalling molecules, postsynaptic scaffold modules and regulatory proteins. Within the group of PSD scaffolds, genes encoding Shanks have raised much interest, as it was suggested that mutations in SHANK1-3 are found in about 1% of all patients affected by ASD2,10,11.
Intriguingly, genetic variants of DLGAP1-4, encoding SAPAP1-4, have been linked with different neuropsychiatric conditions, including ASD, thus implicating these PSD scaffold proteins in the etiology of these disorders9,12,13,14,15,16,17,18,19. After birth, Dlgap1-4 are broadly expressed throughout the rodent brain with some regional differences20,21. While the structural homology between SAPAPs implies some degree of functional redundancy22, it is currently unclear whether they perform similar tasks at different synapses or have evolved to assemble functionally distinct synaptic signalling complexes operating in parallel at single synapses. First clues to these issues are emerging from knockout mouse studies. Thus, SAPAP3-deficient mice exhibit a phenotype reminiscent of obsessive-compulsive disorder (OCD) in humans that could be attributed to the specific expression of SAPAP3 in the striatum23 and an enhanced synaptic signalling mediated via mGluRs24,25. In contrast, the loss of SAPAP2 enhances social dominance and aggressive behaviourFull size image
Heterozygous Dlgap4+/geo and homozygous Dlgap4geo/geo males and females are viable and fertile. Homozygous Dlgap4geo/geo animal frequency for offspring from heterozygous breeding pairs was slightly lower than the expected Mendelian ratio (19% Dlgap4geo/geo; 55% Dlgap4+/geo; 26% Dlgap4+/+; n = 450), while the sex distribution was nearly balanced (48% males and 52% females).
Loss of SAPAP4 affects dendritic arborisation as well as structure and function of excitatory synapses
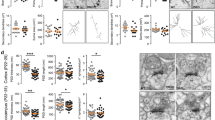
Dlgap4geo/geo animals displayed a normal overall appearance and no alterations were observed with respect to all investigated gross morphological parameters. In particular, overall brain structure of Dlgap4geo/geo mice was normal (Fig. 1e). To assess whether the loss of SAPAP4 alters dendritic arborisation or the shape/density of dendritic spines, we cross-bred Dlgap4geo/geo mice with thy1-GFP line M animals exhibiting sparse enhanced green fluorescent protein (EGFP) labelling of hippocampal pyramidal neurons30 (Fig. 2a). Confocal 3D image stacks were used to reconstruct complete EGFP labelled CA1 neurons (Fig. 2b) or dendritic segments (Fig. 2c)31,32,33. Thus, we found that in the brain of Dlgap4geo/geo mice both the mean total number of branching points (Fig. 2d) and mean total path length of apical dendrites (Fig. 2e) are significantly decreased compared to Dlgap4+/+ animals. For basal dendrites, both parameters are slightly but not significantly reduced (mean total number of branching points, Dlgap4geo/geo: 16.0 ± 2.73, Dlgap4+/+: 19.13 ± 4.73, p < 0.128; mean total path length, Dlgap4geo/geo: 1711.78 ± 310.96 µm, Dlgap4+/+: 1989.43 ± 313.26; p < 0.097; Fisher’s exact test, n = 8 per genotype). In accordance with this finding, Sholl analysis showed a significantly diminished mean number of apical dendritic intersections at 150 and 240 µm distance from the soma in Dlgap4geo/geo mice (Fig. 2f), while it did not reveal significant differences for basal dendritic trees (data not shown). Analysis of dendritic spine morphology further revealed a slight but significant decrease in the fraction of stubby spines in Dlgap4geo/geo mice compared to Dlgap4+/+ animals, while the relative amounts of mushroom-type and thin spines increased to some extent but not significantly (Fig. 2g). The mean spine density remained unchanged (Fig. 2h). Synaptic ultrastructure was further analysed by electron microscopy (EM). We sampled micrographs from randomly chosen areas of the hippocampal CA1 stratum radiatum of Dlgap4geo/geo and Dlgap4+/+ mice. In general, the basic structure of excitatory synapses in the Dlgap4geo/geo brain appeared to be intact. Synapses possessed normally shaped presynaptic boutons containing numerous synaptic vesicles and a dendritic spine containing a clearly visible PSD that was tightly associated with the postsynaptic membrane (Fig. 2i). While the mean area covered by the PSD was increased by 70% in Dlgap4geo/geo compared to Dlgap4+/+ animals (Fig. 2j), the mean density of both excitatory synapses (Fig. 2k) and synaptic vesicles were unchanged (Fig. 2l). The penultimate result is in agreement with our above finding that the mean spine density along dendritic shafts of EGFP labelled pyramidal neurons is identical in Dlgap4geo/geo and Dlgap4+/+ mice (Fig. 2h). To determine if the loss of SAPAP4 may also change the molecular framework of the PSD, we compared the levels of different postsynaptic proteins in PSD enriched fractions derived from hippocampi of Dlgap4geo/geo and Dlgap4+/+ animals. Equal amounts of protein were analysed by Western blotting with antibodies specific for single PSD components. By focussing mainly on scaffold proteins and receptor subunits, we observed that the in vivo loss of SAPAP4 does not noticeably alter the molecular composition of the PSD (Fig. 2m). Noteworthy, we did not observe a compensatory increase in the level of other family members. By trend, postsynaptic levels of the NMDAR subunit GluN1 were found to be enlarged. Taken together, our findings show that SAPAP4 plays an important role in regulating PSD size, while exhibiting only minor effects on spine density and shape.
a Fluorescent micrograph of the hippocampal area obtained from a brain tissue slice of a Dlgap4geo/geo mouse showing EGFP labelling of pyramidal and granule neurons (green). The position of the CA1 region is indicated. Scale bar: 100 μm. b 3D reconstruction of representative CA1 pyramidal neurons showing complete apical (red) and basal (blue) dendritic trees. c Representative images of dendritic segments from hippocampal CA1 pyramidal cells. Scale bar: 10 μm. d, e Bar graphs showing the mean total number of apical branching points (d) and mean total apical dendritic path length (e) of CA1 neurons (*p < 0.05, **p < 0.01, Fisher’s exact test; n = 8 neurons/genotype). Simple vertical lines represent SEM. f Sholl analysis plot showing the mean number of intersections between apical dendrites and a series of concentric spheres centered at the soma of CA1 neurons (*p < 0.05, t-test; n = 8 neurons/genotype). Simple vertical lines represent SEM. g, h Bar graphs displaying the fraction of stubby, mushroom-type and thin dendritic spines (g) and the spine density (h) obtained from EGFP labelled pyramidal neurons of the hippocampal CA1 region of Dlgap4geo/geo and Dlgap4+/+ male mice (14–20 weeks old; *p < 0.05, t-test; n = 10 animals/genotype, ≥63 dendritic segments/genotype, ≥3670 spines/genotype). Simple vertical lines represent SEM. i Representative EM micrographs of excitatory synapses from the hippocampal CA1 area of adult Dlgap4+/+ and Dlgap4geo/geo mice. Scale bar: 500 nm. j–l Bar graphs showing the mean PSD size (j *p < 0.05, t-test; n = 3 mice/genotype, ~70 synapses/animal), the mean density of excitatory synapses (k n = 3 mice/genotype, ~13,500 μm2 CA1 area/mouse) and the mean density of synaptic vesicles (l n = 3 mice/genotype, 75 presynaptic boutons/genotype) obtained from EM micrographs of the hippocampal CA1 molecular layer of adult male mice. m Dlgap4geo/geo to Dlgap4+/+ ratio in the levels of distinct PSD components. In PSD enriched fractions obtained from hippocampal lysates of adult mice, individual proteins were quantified by Western blotting (representative examples are shown on the right). In the bar graph (left), a value of 1 indicates that the level of the respective PSD component is identical in Dlgap4geo/geo and Dlgap4+/+ brain fractions. For each protein, data are based on at least three different PSD preparations and three independent Western blot experiments per preparation. Simple vertical lines in (j–m) represent SD. The following abbreviations have been used: GluN: glutamate receptor ionotropic, NMDA; Gria: glutamate receptor ionotropic, AMPA; mGluR5: metabotropic glutamate receptor 5; Actn2: alpha-actinin-2; Akap1: A-kinase anchor protein 1, mitochondrial; Baiap2: brain-specific angiogenesis inhibitor 1-associated protein 2; Dlg: disks large homolog; GKAP: guanylate kinase-associated protein; Grip1: glutamate receptor-interacting protein 1; Magi1: magi1 protein; Nsmf: NMDA receptor synaptonuclear signaling and neuronal migration factor; SAPAP: SAP90/PSD-95-associated protein; Shank: SH3 and multiple ankyrin repeat domains protein; αCaMKII: calcium/calmodulin-dependent protein kinase type II subunit alpha; SynGAP1: Ras/Rap GTPase-activating protein SynGAP1. For further details see text
To analyse whether the dramatic increase in average PSD area leads to alterations in synaptic transmission, we performed extracellular field and whole-cell patch-clamp recordings from CA1 pyramidal neurons in acute hippocampal slices. Input/output curves of field excitatory postsynaptic potentials (fEPSPs) were similar between Dlgap4+/+ and Dlgap4geo/geo animals (Fig. 3a). We observed a significant rise in the average amplitude of miniature excitatory postsynaptic currents (mEPSCs) in SAPAP4-deficient compared to Dlgap4+/+ brains indicating an increased AMPAR response at active synapses (Fig. 3b). However, both average half width and frequency of mEPSCs were not affected by the loss of SAPAP4 (Fig. 3c), showing that the electrophysiological characteristics of synaptic AMPARs are unchanged and that SAPAP4 loss does not overtly affect presynaptic neurotransmitter release. Intracellular recordings of evoked excitatory postsynaptic currents (eEPSCs) utilizing different subtype-specific glutamate receptor inhibitors revealed that in CA1 pyramidal cells the relative amount of GluN2A-mediated NMDAR currents is significantly decreased in Dlgap4geo/geo mice (Fig. 3d), indicating that SAPAP4 promotes the early postnatal switch from mostly GluN2B- to more GluN2A-containing NMDAR in the hippocampus34. To assess how these changes in synaptic transmission influence the plasticity of synapses, we tested the ability to induce long-term depression (LTD) and long-term potentiation (LTP) at hippocampal CA1 pyramidal cell synapses. While LTD induced by low-frequency stimulation was impaired in Dlgap4geo/geo hippocampus (Fig. 3e), LTP induced by repetitive thetaburst stimulation of the Schaffer collaterals remained intact (Fig. 3f). To assess if SAPAP4 deficiency may affect the maturation of global in vivo neuronal network activity, we monitored discontinuous oscillatory activity of neuronal networks in the prefrontal cortex (PFC) and hippocampus by performing multi-site recordings of local field potentials (LFP) and spiking activity from neonatal mice35,36. Here, we found that in both brain regions the amplitude of oscillatory events was significantly increased in vivo in Dlgap4geo/geo animals compared to Dlgap4+/+ mice, while there was a significant decline in their duration (Fig. 3g). The occurrence of oscillatory events in both brain areas was unchanged. In summary, these findings indicate that SAPAP4 may play a role in the control of AMPAR-mediated synaptic strength, maturation of excitatory synapses and functioning of neuronal circuits.
a Input/output curves of fEPSPs in CA1 stratum radiatum are indistinguishable between Dlgap4+/+ and SAPAP4-deficient littermates. b AMPAR-mediated mEPSCs are significantly larger in SAPAP4-deficient mice. Upper panel: raw traces recorded in exemplary neurons presented at low (upper traces, scale bars 1 s and 20 pA) and at high resolution (lower traces, bars are 10 ms and 10 pA). Lower left panel: cumulative histogram of mEPSC amplitudes shows a significant right-shift in SAPAP4-deficient neurons (**p < 0.01, 3600 events collected from 12 Dlgap4geo/geo cells and 4150 events from 14 Dlgap4+/+ cells, Kolmogorov–Smirnov 2 sample test). Lower right panel: the mean mEPSC amplitude is significantly enhanced in SAPAP4-deficient neurons (**p < 0.01, t-test; n = 12 Dlgap4geo/geo and 14 Dlgap4+/+ cells). c mEPSC frequency (left bar graph) and half-width (right) were indistinguishable between Dlgap4geo/geo and Dlgap4+/+ neurons. d The GluN2A component of the NMDAR-mediated eEPSC is significantly decreased in SAPAP4-deficient neurons (**p < 0.05, t-test; n = 10 Dlgap4geo/geo and 9 Dlgap4+/+). Left panel: exemplary traces of NMDAR-mediated eEPSCs at +40 mV in the presence of CNQX and after application of the GluN2B-antagonist RO 25-6981. Scale bars: 5 ms and 30 pA for Dlgap+/+ and 5 ms and 20 pA for Dlgapgeo/geo eEPSCs. e Following LFS, SAPAP4-deficient slices (n = 14 slices, 4 mice) exhibit reduced LTD compared with Dlgap4+/+ slices (n = 14 slices, 4 mice). Two-way ANOVA revealed significant effects of genotype (F1,14 = 59.2, ***p < 0.001) and time (F1,14 = 2.1, *p < 0.05) but no interaction between these factors (F1,14 = 0.02, p > 0.05). f LTP induced by repetitive thetaburst stimulation (4× TBS) is identical in SAPAP4-deficient (n = 30 slices, 8 mice) and Dlgap4+/+ (n = 30 slices, 7 mice) animals (circles and triangles represent stimulated and control pathways, respectively). Two-way ANOVA revealed a significant effect of time (F1,30 = 7.48, *p < 0.05) but no significant effect of genotype (F1,30 = 2.13, p > 0.05) or an interaction between them (F1,30 = 0.17, p > 0.05). Experiments shown in (a–f) were performed with 12–16-week-old male mice. g Discontinuous oscillatory in vivo activity of prefrontal (PFC) and hippocampal (HC) networks of neonatal (P8–12) mice (8 animals/genotype). The bar graphs illustrate that in both investigated brain areas the amplitude of oscillatory events was significantly amplified in Dlgap4geo/geo mice (p < 0.001, two-sample Kolmogorov–Smirnov test), whereas their duration was shortened (p < 0.001). SAPAP4 deficiency did not alter the occurrence of oscillatory events in both brain regions (p = 0.99). Simple vertical lines represent SEM. For more details see text
Dlgap4 geo/geo mice exhibit hyper-locomotion and disrupted novelty-induced exploratory behaviour
As genetic variants of DLGAP1-4 have been linked to abnormal behaviour in humans9,12,13,14,15,16,17,18,19, we performed a variety of behavioural tests with littermates from heterozygous breeding pairs. Dlgap4 ablation profoundly altered novelty-induced exploratory behaviour as Dlgap4geo/geo mice showed hyper-locomotion indicated by the longer distance moved in the open field test (Fig. 4a). Noteworthy, Dlgap4geo/geo mice did not display any short-term (‘within-session’) habituation. While the distance moved by Dlgap+/+ mice diminished as the trial progressed, the mean distance travelled by Dlgap4geo/geo mice remained constant (Fig. 4a). Also, the number of rearing bouts, a typical exploratory behaviour in rodents37, was significantly reduced in SAPAP4-deficient mice compared to Dlgap4+/+ littermates (Fig. 4c), whereas thigmotactic behaviour was not affected (Fig. 4b). These observations show that Dlgap4geo/geo mice exhibit severe hyper-locomotion and impaired habituation learning, while reduced rearing indicates that novelty-induced exploratory behaviour may be disturbed.
a Dlgap4geo/geo mice covered longer distances in the open field test as compared to Dlgap4+/+ mice (***p < 0.001, effect of genotype after mixed two-way ANOVA). b Thigmotactic behaviour in the open field was not affected in Dlgap4geo/geo mice. c Rearing, a typical novelty-induced exploratory behaviour in mice, was reduced in Dlgap4geo/geo mice during the first 5 min of the open field compared to Dlgap4+/+ mice. d–h In the elevated plus maze test, Dlgap4geo/geo mice spent more time on the open arms (d), entered the open arms more often (e), did more total transitions (f), spent less time on the centre (g) and self-groomed less (h) compared to Dlgap4+/+ mice (in (c–h): *p < 0.05, **p < 0.01 and ***p < 0.001; unpaired t-test). i During the dark cycle, Dlgap4geo/geo mice in the home cage were more active than Dlgap4+/+ mice. (**p < 0.01 as compared to Dlgap4+/+ mice within the time bin, Newman–Keuls post-hoc test after mixed three-way ANOVA). Simple vertical lines represent SEM. All experiments were performed with 11 adult male mice/genotype (cohort 1). For more details see text
In the elevated plus maze (EPM) test, significantly more Dlgap4geo/geo mice fell from the open arm as compared to Dlgap4+/+ littermates (Dlgap4geo/geo: 5/11, Dlgap4+/+: 0/11; p < 0.05, Fisher’s exact test). Also, ambulation of Dlgap4geo/geo animals lacked the typical ‘flat’ posture displayed by mice walking on an elevated open arm for the first time (data not shown). Both observations imply that Dlgap4geo/geo mice display impaired risk-assessment, lack of attention, sensory-motor deficits, or a combination of these factors. Behavioural analysis of all mice that completed the whole 5 min EPM test phase (n = 6 Dlgap4geo/geo and 11 Dlgap4+/+ males) revealed a high preference of Dlgap4geo/geo animals for exploring open versus closed arms (Fig. 4d, e), indicative of a decrease in anxiety. Also, Dlgap4geo/geo mice displayed an enhanced general locomotion as indicated by the increased number of transitions during the test period (Fig. 4f), consistent with the hyperactivity displayed in the open field test. Furthermore, Dlgap4geo/geo animals spent less time in the centre of the maze (Fig. 4g), a position in which mice normally remain to assess potential risks of entering the open arms, thus again indicating a disrupted risk-assessment. Finally, similar to the open field test the number of rearing bouts was reduced (Dlgap4geo/geo: 3.0 ± 1.3, Dlgap4+/+: 14.5 ± 1.1; p < 0.001, Fisher’s exact test), further supporting an impaired novelty-induced exploratory behaviour. Given the OCD-like behaviour of Dlgap3−/− mice23, we also monitored self-grooming. Up to 1-year-old Dlgap4geo/geo animals did not display any signs of skin lesions in head, neck or snout regions that may have arisen from excessive self-grooming (data not shown). During the EPM test phase, self-grooming was only displayed by one out of six tested Dlgap4geo/geo mice. As all tested Dlgap4+/+ littermates (n = 11) engaged in self-grooming, its total time was significantly reduced in Dlgap4geo/geo animals (Fig. 4h; p < 0.01, Fisher’s exact test). Thus, Dlgap4geo/geo mice spent less time in a posture indicative of a conflictual state and arousal38.
When kept in their home cages, Dlgap4geo/geo mice displayed an increased locomotor activity during the dark phase (Fig. 4i; p < 0.01, Newman–Keuls post-hoc test) that was similar to the enhanced activity observed in the open field and EPM test. The general circadian activity profile however was unaltered. Thus, hyper-locomotion of Dlgap4geo/geo mice affects both home cage activity and adaptive behavioural responses.
Cognitive functioning is disrupted in Dlgap4 geo/geo mice
To assess cognitive function, we performed the spontaneous alternation test. It relies on the preference of mice to rather explore unfamiliar than familiar stimuli. Given the choice to enter two different arms of a Y-shaped maze, freely moving animals will thus choose to enter the less recently visited arm. To successfully perform this task, animals need to remember the order in which the three arms of the maze have been visited, a function requiring working memory. While Dlgap4+/+ mice showed the expected preference for the less recently visited arm during both sessions performed on days 1 and 2 (Fig. 5a), Dlgap4geo/geo mice alternated at chance level (50%) potentially suggesting working memory impairment. However, this poor performance may as well result from their hyperactive behaviour and a consequential decreased attention, an assumption that is supported by the fact that Dlgap4geo/geo mice moved faster between arms than Dlgap4+/+ animals (Fig. 5b). Thus, we investigated spatial learning and memory in the water maze test, an experiment that does not involve novelty-seeking as the spontaneous alternation test. Spatial learning and memory was significantly impaired in Dlgap4geo/geo mice as indicated by their poorer performance during learning (Fig. 5c) as well as by their searching strategy during the probe trial (Fig. 5d–f). During learning, they required more time to find the hidden platform (Fig. 5c) compared to Dlgap4+/+ mice, despite a similar swim velocity (data not shown). During the probe trial, Dlgap4geo/geo mice spent equal amounts of time in all four quadrants with no preference for the target quadrant (Fig. 5d). Also, they searched at larger mean distance to the former platform position compared to Dlgap4+/+ mice (Fig. 5e). These data indicate that Dlgap4geo/geo mice were not able to locate the hidden platform and most likely only found it based on an alternative searching strategy such as circling. Therefore, SAPAP4-deficient animals use simple information as for example the approximate distance of the platform from the pool wall (Fig. 5f). Taken together, data obtained with the spontaneous alternation and water maze test suggest that SAPAP4 deficiency leads to cognitive deficiencies resulting from impaired spatial learning, attention deficits or a combination of both.
a During both days of the spontaneous alternation test, Dlgap4+/+ mice exhibited higher percentages of alternations than expected by chance (50%, indicated by dotted line; ##p < 0.01, Wilcoxon signed rank test) and as compared to Dlgap4geo/geo mice (*p < 0.5 and **p < 0.01, Mann–Whitney test). b The average time to perform a transition increased from day 1 and day 2 in Dlgap4+/+, but not Dlgap4geo/geo mice (§§§p < 0.001 Newman–Keuls post-hoc test after mixed two-way ANOVA) and was lower in Dlgap4geo/geo compared to Dlgap4+/+ mice (**p < 0.01 and ***p < 0.001, Newman–Keuls post-hoc test after mixed two-way ANOVA). c Impaired spatial learning in Dlgap4geo/geo mice as indicated by the increased escape latencies during the learning trials of the water maze test (***p < 0.001, effect of genotype after mixed two-way ANOVA). d–f During the probe trial of the water maze test, Dlgap4geo/geo mice spent less time in the target quadrant (d ***p < 0.001, Newman–Keuls post-hoc test after mixed two-way ANOVA) and swam at longer distances from the platform compared to Dlgap4+/+ mice (e ***p < 0.001, unpaired t-test). However, mice of both genotypes swam at the appropriate distance from the maze wall in order to find the platform (f the entire area with a platform to wall distance identical to the respective distance during the learning trials is displayed in grey). Simple vertical lines represent SEM. All experiments were performed with 11 adult male mice/genotype (cohort 1). For more details see text
Dlgap4 geo/geo mice exhibit impaired social behaviour and vocal communication
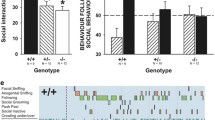
In humans, DLGAP1-4 have been identified as risk genes for different neuropsychiatric disorders39,40. Thus, we tested how Dlgap4geo/geo mice respond to social stimuli. To assess communicative functions, we recorded ultrasonic vocalizations (USVs) of juvenile mice emitted during direct reciprocal social interaction in same-genotype pairs. In this situation, USVs appear to have a pro-social affiliative function and may help to maintain social proximity41. The total number of USVs emitted throughout the entire test session was dramatically reduced in Dlgap4geo/geo mice compared to Dlgap4+/+ animals (Fig. 6a). This was accompanied by a strong decrease in the time Dlgap4geo/geo mice engaged in social interactions (Fig. 6b). The reduction in both parameters was particularly apparent during the first minute of the encounter, when Dlgap4+/+ mice displayed a high interaction rate that gradually faded during the course of the test session.
a Total number of ultrasonic vocalizations emitted during the 5 min social interaction test in juveniles (bar graph), with the time course for the number of ultrasonic vocalizations emitted for each 1 min time bin across the 5 min social interaction period, plus 1 min habituation (line diagram; dashed line indicates introduction of partner mouse; repeated measurements ANOVA with the between-subject factor genotype and the between-subject factor test duration; **p < 0.01 and ***p < 0.001). b Total social interaction time displayed during the 5 min direct reciprocal social interaction test in juveniles (bar graph), with the time course for the social interaction time displayed during each 1 min time bin across the 5 min social interaction period, plus 1 min habituation (line diagram; dashed line indicates introduction of partner mouse; repeated measurements ANOVA with the between-subject factor genotype and the between-subject factor test duration; *p < 0.05, **p < 0.01, ***p < 0.001). Experiments shown in (a and b) were performed with 18 Dlgap4+/+ (9 male and 9 female) and 11 Dlgap4geo/geo mouse pairs (6 male and 5 female) from cohort 2, respectively. c, d In adulthood, Dlgap4geo/geo mice spent less time investigating a male or female conspecific during direct reciprocal social interaction (c). On average, each social approach was significantly shorter as compared to Dlgap4+/+ mice (d) (*p < 0.05, **p < 0.01, ***p < 0.001; Newman–Keuls post-hoc test after mixed two-way ANOVA). Simple vertical lines represent SEM. Experiments shown in (c and d) were performed with 10 adult male focal mice/genotype (cohort 3). For more details see text
The above social interaction assay requires the use of same-genotype/-sex pairs as recorded USVs cannot be attributed to an individual mouse. Thus, we next explored direct reciprocal social interactions between adult Dlgap4geo/geo and Dlgap4+/+ mice. Here, an unfamiliar age- and body-weight-matched male mouse (intruder) was introduced for 10 min in the home cage of the male focal animal (resident). As no aggressive interaction was observed between residents and intruders, our behavioural analysis was restricted to the following parameters: time spent sniffing the intruder, allo-grooming, digging and self-grooming. Whereas no differences between genotypes were detected for allo-grooming, digging and self-grooming, Dlgap4geo/geo mice spent significant less time sniffing the intruder compared to Dlgap4+/+ mice (Fig. 6c), which was not due to a lower amount of approaches, but to the fact that on average each approach lasted longer in Dlgap4+/+ mice compared to Dlgap4geo/geo mice (Fig. 6d). These findings indicate that SAPAP4-deficient mice have a reduced ability for maintaining social interactions. Similar results were obtained when we used an unfamiliar adult female mouse as intruder (Fig. 6c, d). Thus, Dlgap4geo/geo mice display severe deficits in social interaction independent of whether they encounter SAPAP4-deficient or Dlgap4+/+ mice.