Abstract
Bronchus-associated lymphoid tissue (BALT) develops at unpredictable locations around lung bronchi following pulmonary inflammation. The formation and composition of BALT have primarily been investigated by immunohistology that, due to the size of the invested organ, is usually restricted to a limited number of histological sections. To assess the entire BALT of the lung, other approaches are urgently needed. Here, we introduce a novel light sheet microscopy-based approach for assessing lymphoid tissue in the lung. Using antibody staining of whole lung lobes and optical clearing by organic solvents, we present a method that allows in-depth visualization of the entire bronchial tree, the lymphatic vasculature and the immune cell composition of the induced BALT. Furthermore, three-dimensional analysis of the entire lung allows the qualitative and quantitative enumeration of the induced BALT. Using this approach, we show that a single intranasal application of the replication-deficient poxvirus MVA induces BALT that constitutes up to 8% of the entire lung volume in mice deficient in CCR7, in contrast to wild type mice (WT). Furthermore, BALT induced by heat-inactivated E. coli is dominated by a pronounced T cell infiltration in Cxcr5-deficient mice, in contrast to WT mice.
Similar content being viewed by others
Introduction
Bronchus-associated lymphoid tissue (BALT) was originally described as clusters of lymphoid tissue embedded along the walls of large airways in the lung.1 Unlike other secondary lymphoid tissues, BALT is not constitutively present in all species2 but can be induced by exposure to microbes or other compounds, leading to lung inflammation.3,4,5,6 As a lymphoid tissue, BALT functions in the induction of protective host immune responses through the recruitment, priming and proliferation of antigen-specific lymphocytes.4,7,8 To further understand the functions of induced BALT, it is necessary to study its development and organization in response to diverse stimuli. Immunohistology of lung cryosections is often used as a standard method to determine the development and organization of BALT in different animal models. In this technique, representative lung cryosections are prepared, stained with antibodies to label BALT and imaged using a fluorescence microscope to visualize and quantify the amount of BALT in the sections. However, this method has several limitations. It is labor-intensive and difficult to assure quality standards, particularly as the distribution of induced BALT within individual lung lobes may vary significantly from animal to animal.
The advancement of light sheet microscopy (LiSM) has improved the study of structures within large tissues or whole organs in three dimensions (3D). During LiSM imaging, excitation light is focused by a cylindrical lens onto a sheet of light that illuminates only the focal plane of the detection optics.9 The specimen is illuminated by a thin sheet of light, horizontally, to reduce photobleaching and out of focus blur;10 in the process of imaging, several single image stacks from different excited regions are collected and can be reconstructed into a 3D image of the imaged specimen. A major challenge for imaging whole specimens is that light is directed through cellular and extracellular structures, which have different refractive indices (RIs), causing light scattering that results in images with low resolution and imaging depth.11 Normalization of RIs can be achieved through optical clearing of specimens prior to imaging. A number of optical clearing protocols can be used for imaging specimens from transgenic animals expressing specific fluorescent proteins, as well as antibody-stained specimens.12,13,14,15
In this study, we investigated the application of LiSM for qualitative and quantitative analysis of BALT in whole mouse lung lobes. BALT was identified by lymphocyte-specific antibody staining of whole lung lobes using a protocol similar to ‘immunolabeling-enabled three-dimensional imaging of solvent cleared organs’ (iDISCO)16 and optically cleared using an organic solvent-based protocol, three-dimensional imaging of solvent-cleared organs (3DISCO),13 prior to LiSM analysis. In addition, we investigated the induction of BALT by heat-inactivated Escherichia coli and characterized its development and organization using immunohistology and LiSM.
Materials and methods
Mice
All mice used in our experiments were bred locally at the animal facility of the Hannover Medical School under specific pathogen-free conditions. Ccr7 −/−, Cxcr5 −/−, Myd88/Trif −/− , Myd88/Trif/Cardif −/−, and Ifnαr1 −/− mice have been described previously.17,18,19,20,21,22 All experiments were performed in accordance with local animal welfare regulations reviewed by the institutional review board and the Lower Saxony State Office for consumer protection and food safety.
MVA and BALT induction
Recombinant modified vaccinia virus Ankara (MVA) viruses have been described previously and were generated by standard methods.4,5,23 In brief, the virus was propagated in chicken embryo fibroblasts, concentrated and purified by ultracentrifugation through sucrose. To determine the infectious units (IU) for use, viruses were titrated in chicken embryo fibroblasts. Before virus administration, mice were deeply anesthetized with a mixture of Ketamine (Albrecht GmbH) and Xylazine (Bayer Vital GmbH). MVA was diluted to a final concentration of 3.3 × 108 IU/ml in PBS (Biochrom GmbH), and 30 μl (107 IU) was administered intranasally.
E. coli
E. coli, strain DH5-α, was cultivated in Luria-Bertani (LB) medium (BD). Bacteria were plated on LB-agar (Carl Roth) and incubated overnight at 37 °C. A single colony culture was prepared in 10 ml of LB broth (Carl Roth) and incubated as above, on a shaker. The bacterial suspension was diluted 1:200 in 100 ml LB broth and incubated as above. Bacteria were harvested at an optical density of 600 (OD600), pelleted, washed and re-suspended in PBS. Colony forming units (CFU) were determined by plating different dilutions and counting using a Thoma cell counting chamber (Brand GmbH); thereafter, they were heat-inactivated (30 min at 50 °C and an additional 15 min at 85 °C) in a water bath. Heat-inactivation was confirmed by plating bacteria on LB-agar and Columbia-blood-agar (Oxoid) and incubating for 72 h. For induction of BALT, anesthetized mice were administered a single 30 μl suspension of 3 × 108 CFU, as determined by titration, intranasally.
Immunohistology
Lung cryosections were stained with anti-CD3-Cy3 (17A2), anti-B220-AlexaFluor647 (RA3-GB2) (all prepared in-house) and anti-CD21/35-FITC (7G6; BD) antibodies. For chemokine staining, goat anti-CXCL12 (polyclonal, PeproTech) and goat anti-CXCL13 (R&D Systems) antibodies were detected with donkey anti-goat HRP (Jackson ImmunoResearch) and amplified with a Tyramide Signal Amplification system (PerkinElmer). Imaging was performed on an epifluorescence microscope equipped with an F-View II camera (Olympus). The numbers of BALT structures per section were counted using the polygon tool in cellSens software (Olympus).
Lymphocyte labeling
Lymphocytes isolated from wild-type (WT) mouse spleens were labeled with TAMRA (Invitrogen) as previously described.24 Briefly, re-suspended lymphocytes were pre-warmed for 15 min at 37 °C in a water bath, mixed with TAMRA and incubated for another 12 min. Labeling effectiveness was checked by flow cytometry analysis. TAMRA-labeled lymphocytes were transferred intravenously into BALT-bearing mice via the tail vein. Mice were sacrificed 18 h later, and lungs were collected and analyzed by LiSM.
Antibody staining of entire lung lobes
Mice were sacrificed, and the lungs were perfused with 10 ml of cold PBS through the heart; they were then filled with 1 ml of 4% PFA (MP Biomedicals) through the trachea to prevent the lungs from collapsing and significantly changing their volume during the optical clearing process. Lungs were carefully removed and incubated for 2 h in 3 ml of 4% PFA for further fixation. Lung lobes were separated and washed twice in PBS and then incubated overnight in a blocking/permeabilization buffer consisting of PBS, 0.1% Tween 20 (Carl Roth) and 10% rat serum (Innovative Research). Lung lobes were then incubated for 3 days with 2 μg/ml anti-CD3-Cy3 (17A2) and 2 μg/ml anti-B220-AlexaFluor647 (RA3-GB2) to stain T and B cell aggregates within BALT, along with 1 μg/ml anti-Thy1.2-SeTau647 (MMT1) to stain lymphatic vessels. All antibodies were prepared in-house, and dilutions were prepared in PBS with 0.05% Tween20.
Lung lobe optical clearing and LiSM
Clearing of lung lobes was performed based on the 3DISCO protocol described previously.13 In brief, lung lobes were incubated for 30 min in 50, 70 and 80% (all vol/vol diluted in dH2O)Tetrahydrofuran (THF; Sigma Aldrich), followed by three 30 min incubations in 100% THF, and finally, incubation in Dibenzyl ether (DBE) (Sigma-Aldrich). Lobes were imaged in DBE, using an UltraMicroscope II (LaVision BioTec). During imaging, specimens were excited with a 488-nm laser for detection of autofluorescence; for detection of Cy3- and Alexa647-conjugated antibodies, samples were excited with 561-nm and 647-nm lasers, respectively. A step size of 10 μm was used to collect images of the whole lung lobes, while for smaller regions of interest, a step size of 5 μm was used. Image analysis was performed using Imaris software (Bitplane). TIFF files collected from the light sheet microscope were reconstructed into representative 3D images. Using the Surfaces tool of the Imaris software, the lung lobe and lymphoid aggregate volumes were determined separately, based on thresholds corresponding to their respective fluorescence intensities.
Statistics
The analysis of measurement variables and graphical presentations were performed using GraphPadPrism 5 software (GraphPad Software, Inc). Comparisons within groups were determined by either unpaired two-tailed tests (Mann-Whitney tests) or One-Way ANOVA (Kruskal-Wallis tests). Differences were considered statistically significant for P-values<0.05.
Results
Sufficient optical clearing of lung lobes using the 3DISCO protocol for LiSM
The main limitation of imaging whole organs is scattering of the light when it passes through structures with different RIs.11 Light scattering leads to losses in excitation and emission efficiency, resulting in images with poor resolution and low imaging depth.25 Optical clearing of organs to match the RIs within tissues and the imaging solvent is the only way to overcome this limitation. Several protocols for optical clearing of organs for microscopy analysis have been reported.12,13,14,15 In our first experiment, we determined the optimal clearing protocol for the lung.
First, we tested the clear lipid-exchange Acrylamide-hybridized imaging tissue hydrogel (CLARITY) method, which is an aqueous solution-based clearing protocol that has been used for visualization of transgenic immunofluorescent proteins and antibodies.15 We used a simplified version of the protocol that was optimized for visualization of antibody-stained brain specimens in rats.26 Lungs from WT mice were perfused with a paraformaldehyde/hydrogel solution and incubated overnight to allow complete tissue polymerization. Polymerized lobes were optically cleared in Boric acid and imaged on a light sheet microscope in 2, 20-Thiodethanol. We noticed that the clearing depth was low, and only the outer regions of the lung could be cleared and imaged. Furthermore, the lung lobes remained fragile, making it difficult to handle the samples for imaging of their entire volume (data not shown).
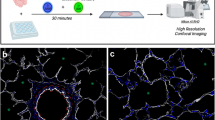
Next, we tested an organic solvent-based clearing protocol, 3DISCO.13 This protocol is faster and better suited for clearing antibody-stained tissues.27 Lung lobes from WT mice were dehydrated and cleared with THF and DBE, after which they were completely transparent (Figure 1a). Solvent-based clearing of whole organs is associated with a reduction of their volume.28 Even though lungs were fixed to retain their physiological shape, the lobe volume reduced noticeably after clearing. Cleared lobes were imaged by LiSM, where they were illuminated horizontally with a thin light sheet (Figure 1b) to reduce the generation of out-of-focus light that would lead to low-quality images. Z-stacks for entire lobes were collected and analyzed using Imaris software. From these images, it was possible to view individual optical sections across the entire lobes (Figure 1c), as well as their 3D reconstructions (Figure 1d and Supplementary Movie S1). Images had excellent quality with sufficient imaging depth, which allowed visualization of the entire lung anatomy.
3DISCO clearing and light sheet microscopy (LiSM) imaging of whole lung lobes. (a) Clearing of separated lung lobes using organic solvents. Separated lung lobes (left) were dehydrated in increasing concentrations of THF followed by optical clearing in DBE (right) before LiSM imaging. (b) Diagrammatic representation of LiSM set-up showing sample orientation, excitation by a thin light sheet and image acquisition. (c, d) Representative LiSM micrographs from 3DISCO-cleared lung lobes, showing sectional and 3D views of an imaged lobe from a wild type (WT)mouse. AF, autofluorescence.
BALT within lung lobes can be visualized by antibody staining
After successfully using 3DISCO to clear lung lobes for imaging by LiSM, we next examined whether this method could be applied to visualize BALT within them. BALT was induced in mice upon intranasal application of MVA, as described previously,4,5 or formed spontaneously in Ccr7 −/− mice.29 In these models, two different approaches were assessed to label BALT within lobes for visualization using LiSM.
The first approach was to intravenously transfer dye-labeled cells that would home to BALT.4 Lobes from recipient mice were analyzed 18 h later. Transferred cells efficiently homed to the lung and were detected within BALT (Figure 2a and Supplementary Movie S2). However, several TAMRA-labeled cells and cells that ingested dead TAMRA-positive cells, such as macrophages, created nonspecific signals within the lung parenchyma. Even though this approach allowed visualization of BALT, its further analysis would be affected by the degree of nonspecific background signals.
Identification of bronchus-associated lymphoid tissue (BALT) in lung lobes for light sheet microscopy (LiSM) imaging. (a) Representative LiSM micrograph from a lung lobe, showing TAMRA-labelled lymphocytes (red) that were intravenously transferred into BALT-bearing mice before LiSM imaging to identify BALT. (b, c) Representative LiSM images showing lymphocyte-specific antibody staining to visualize BALT within lung lobes. Whole-lung lobes from a Ccr7 −/− mouse or WT mouse (as a control) were stained with anti-CD3 (red) and anti-B220 (blue) to identify and visualize BALT following LiSM imaging. (d) Representative LiSM micrograph showing Thy1 staining (blue) in a lung lobe from an untreated wild type (WT)mouse. AF, autofluorescence.
The second approach was to label BALT with lymphocyte-specific antibodies. This was done using a protocol modified from one previously described by Reiner et al. called iDISCO, which has been used to visualize structures and processes within different organs.16 Because BALT is mainly composed of T and B cells, we stained lung lobes with anti-CD3 and anti-B220 antibodies before clearing and LiSM. Directly labeled antibodies conjugated with Alexa Fluor compared to staining with Cyanide dyes resulted in lower background signals, especially for higher-spectrum wavelengths (data not shown). An overnight incubation in PBS-serum supplemented with Tween20 was optimal to facilitate antibody penetration, and lymphocytes within BALT were successfully stained after 72 h (Figure 2b and Supplementary Movie S3). Antibody staining was highly specific for lymphoid aggregates, with no to very little nonspecific background staining. As expected, no lymphoid aggregate staining was detected in lung lobes from WT control mice (Figure 2c and Supplementary Movie S4). These data demonstrate that antibody staining of lymphocytes within lung lobes was adequate for visualization of BALT following LiSM. Interestingly, staining with an anti-Thy1 mAb allowed the visualization of the entire lymphatic vessel tree within the lung (Figure 2d).
BALT within lung lobes could be quantified following LiSM
We next quantified BALT within LiSM-imaged lung lobes using Imaris software’s ‘Surfaces’ tool by rendering the volume surfaces of the entire lung lobe, as well as individual T cell and B cell aggregates (Figure 3a and Supplementary Movie S5). In both, WT and Ccr7 −/− murine B and T cells contributed equally to BALT (Figure 3b). As expected, there was no BALT in lobes from untreated WT mice, except for a few isolated lymphoid aggregates, whereas these structures were readily present in lung lobes from untreated Ccr7 −/− mice (Figure 3c). We determined the volume of the left lobe, right inferior lobe, right superior lobe and right middle lobe. The right post-caval lobe was not included in the analysis because of its substantial reduction in volume following clearing, leading to technical challenges during imaging. The volume of individual lung lobes was calculated as the percentage of the sum of the 4 lobes analyzed. No differences in the distribution of the whole lobe volumes between WT and Ccr7 −/− mice were detectable (Figure 3d).
Quantification of bronchus-associated lymphoid tissue (BALT) in light sheet microscopy (LiSM)-imaged lung lobes. (a) Surface renderings of antibody-stained and LiSM-imaged lung lobes isolated from a Ccr7 −/− mouse, using Imaris software, to determine the volume of T-cell (anti-CD3, red) and B-cell (anti-B220, blue) aggregates, which were subsequently used to calculate the volume of BALT within lobes. (b) Percent volume of T- and B-cell aggregates per lung lobe quantified in WT (left) and Ccr7 −/− (right) mice. (c) Percent volume of BALT per lung lobe quantification in WT and Ccr7 −/− mice. (d) Relative volume of whole lung lobes from WT and Ccr7 −/− mice. Individual lung lobe volume was calculated as the percent of the sum of left, right inferior, right superior and right middle lobes. For quantitative analysis, eight mice were included per group, with at least six independent experiments performed. Bars represent median values; ***P<0.001. AF=autofluorescence. Scale bars=500 μm.
Qualitative and quantitative analyses of BALT with LiSM and immunohistology are similar
Next, we analyzed MVA-induced BALT in WT and Ccr7 −/− mice by LiSM and compared the results to immunohistology analyses in which lung cryosections were stained with DAPI, to identify cellular aggregates. As expected, aggregates could not be found in cryosections from uninfected WT mice but were found following MVA infection. In contrast, BALT could be found in cryosections from uninfected Ccr7 −/− mice and considerably increased in size and number following MVA-infection (Figures 4a and b). Immunohistological observations were similar to LiSM, with lobes stained with antibodies specific for B and T cells. There was no staining in the lung lobes from uninfected WT mice, while prominent staining was detected after MVA-infection (Supplementary Movie S6).
Comparing qualitative and quantitative analysis of modified vaccinia virus Ankara (MVA)-induced bronchus-associated lymphoid tissue (BALT) between immunohistology and light sheet microscopy (LiSM). (a) Representative immunofluorescence micrographs of WT and Ccr7 −/− lung cryosections from uninfected or MVA-infected mice (12 days post-infection), stained with DAPI (red). (b) Representative LiSM micrographs of WT and Ccr7 −/− lung lobes from uninfected or MVA-infected mice (12 days post-infection) stained with anti-CD3 (red) and anti-B220 (blue) antibodies. (c) Quantitative analysis of the amount of BALT per lung (mean value of 4 sections per lung) from uninfected or MVA-infected WT and Ccr7 −/− mice (WT, uninfected n=6, MVA-infected n=8; Ccr7 −/−, uninfected n=8, MVA-infected n=7). (d) Quantitative analysis, using Imaris software, of the percent volume of BALT per lung (mean value of left lobe, right inferior lobe, right superior lobe and right middle lobe per lung) from uninfected or MVA-infected WT and Ccr7 −/− mice (WT, uninfected n=8, MVA-infected n=9; Ccr7 −/−, uninfected n=8, MVA-infected n=8). For quantitative analysis, at least 6 independent experiments were performed. Bars represent median values; **P<0.01; ***P<0.001. AF=autofluorescence. Scale bars=500 μm.
For immunohistology-based BALT quantification, cryosections were stained with anti-CD3 and anti-B220 antibodies, and individual structures were counted and measured. Four cryosections from around the main bronchi and vessels were analyzed per lung.5,29 There was significantly more BALT per cryosection from MVA-infected WT mice compared to uninfected mice. In Ccr7 −/− mice, as expected, BALT was already present in untreated lungs; however, the amount increased significantly in the MVA-infected group (Figure 4c). Likewise, LiSM revealed a significant increase in the volume of BALT within MVA-infected WT mice compared to uninfected mice, with 0.6% of the total lung being occupied by BALT, on average. In non-infected Ccr7 −/− mice, 0.4% of the total lung volume was occupied by spontaneous BALT on average; this value increased to 4.2% in the MVA-infected group (Figure 4d).
Characterization of E. coli-induced BALT
Different pathogens can induce BALT formation by triggering different molecular pathways.5 E. coli is not a typical lung pathogen but rather a gut commensal; however, there is evidence that it can cause pneumonia, particularly in newborns and patients requiring intensive care.30,31,32,33 Furthermore, an intranasal administration of an E. coli endotoxin, lipopolysaccharide (LPS), induces airway inflammation.34,35,36 We, therefore, investigated whether intranasal administration of a heat-inactivated lab strain of E. coli, DH5-α, could induce the formation of BALT.
WT mice were infected with one dose of E. coli, and lung lobes were examined for BALT formation by LiSM 18 days later. BALT was detected near both the secondary and tertiary bronchi, as well as bronchioles (Figure 5a). At least 0.25% of the E. coli-infected lobes were occupied by BALT (Figure 5b). The distribution and frequency of E. coli-induced BALT on day 18 were similar to that of MVA-induced BALT on day 12, as shown previously,4,5 suggesting that E. coli induces BALT with slower kinetics.
Qualitative and quantitative analysis of E. coli-induced bronchus-associated lymphoid tissue (BALT) in WT mice. (a) Representative light sheet microscopy (LiSM) lung lobe micrograph showing BALT formation following E. coli infection (18 days post-infection). To visualize BALT, lung lobes were stained with anti-CD3 (red) and anti-B220 (blue) antibodies. (b) Quantitative analysis, using Imaris software, of the percentage volume occupied by BALT per lung lobe in LiSM-imaged lung lobes from uninfected and E. coli-infected WT mice. Data were collected from eight uninfected and four E coli-infected mice. Four lung lobes were analyzed per mouse. (c) Representative immunofluorescence micrographs taken over time from lung sections of mice in which BALT was induced by infection with E. coli. BALT was identified by staining cryosections with anti-CD21/35 (green), anti-CD3 (red) and anti-B220 (blue) antibodies. (d) Immunofluorescence micrographs showing how E. coli-induced lymphoid aggregates were classified as either type I, type II or type III based on the presence or absence of organized B cell follicles. Lung cryosections were stained with anti-CD3 (red) and anti-B220 (blue). Quantitative analysis showing (e) relative frequency and (f) size of type I, type II, and type III lymphoid aggregates over time following E. coli infection in wild type (WT)mice. Three mice were analyzed at each time point, and plotted data was collected from 4 cryosections per mouse. Representative immunofluorescence micrographs showing expression levels of (g) CXCL12 and (h) CXCL13 chemokines over time following E. coli infection in WT mice. Error bars (e) represent means±s.d.; bars (b, f) represent median values; *P<0.5; **P<0.01; ***P<0.001. The color of stars (e) relates to the type of lymphoid aggregate, as indicated. AF, autofluorescence.
To determine the cellular composition, frequency and size of E. coli-induced BALT, we examined lung cryosections on days 6, 12, 18 and 30 following induction. After staining with anti-CD21/35 (FDCs), anti-CD3 (T cells) and anti-B220 (B cells) antibodies, the cryosections were analyzed with an epifluorescence microscope. BALT could be observed starting at the earliest time point and consisted of B cell follicles, which became more organized by days 18 and 30, characterized by the presence of follicular dendritic cells (FDCs; Figure 5c).
For further analysis, E. coli-induced lymphoid aggregates were classified into three types according to criteria previously used for the classification of MVA- and P. aeruginosa-induced aggregates.5 Type I aggregates were composed of organized B-cell follicles surrounded by T cells, type II aggregates lacked organized B-cell follicles but contained B and T cells, and type III aggregates predominantly consisted of T cells only (Figure 5d). The relative frequency of type I aggregates was similar between days 6 and 12 but increased significantly on days 18 and 30. In contrast, the opposite trend was observed for type II aggregates, with significantly fewer aggregates found at days 18 and 30; no significant differences were observed for type III aggregates (Figure 5e). In addition to frequency, the size of the individual aggregates was determined. There was no difference in the size of type I lymphoid aggregates from day 6 to day 18; however, they were significantly larger at day 30 compared to day 12 and day 18. Compared to day 6, a significant reduction in the size of type II lymphoid aggregates was observed on days 12, 18 and 30. Likewise, the size of type III aggregates was smaller on days 12 and 18, but their size on day 30 was similar to their size on day 6 (Figure 5f).
Finally, we characterized E. coli-induced BALT by investigating the expression of the C-X-C chemokine ligands 12 and 13 (CXCL12 and CXCL13) over time. Lung cryosections from days 6, 12, 18 and 30 were stained with anti-CD21/35, anti-CXCL12 or anti-CXCL13, and anti-B220 antibodies. While expression of CXCL12 was detected at all time points (Figure 5g), expression of CXCL13 was only observed at days 18 and 30, coinciding with the presence of FDCs. (Figure 5h).
E. coli-induced BALT in Cxcr5 −/− mice is characterized by T-cell aggregates surrounded by B-cell follicles
Since CXCL12 and CXCL13 were expressed in E. coli-induced BALT, we hypothesized that CXCL13/CXCR5 signaling might be involved in the recruitment and organization of B cells. To confirm this hypothesis, we induced E. coli BALT in Cxcr5-deficient (Cxcr5 −/−) mice in order to compare its cellular composition and organization, as well as its relative frequency, with WT mice. Lung cryosections from these mice were analyzed on day 18, which had been seen in previous experiments to be the peak time point for E. coli BALT development. LiSM showed that E. coli was capable of inducing BALT in Cxcr5 −/− mice (Figure 6a), which was similar in volume per lobe to WT mice (Figure 6b). However, when characterizing the ratio of infiltrated T and B cells in the whole lung tissue, lobes of Cxcr5 −/− mice contained significantly more T-cell infiltrates compared to WT mice, whereas B-cell infiltrates were comparable (Figure 6c).
Qualitative and quantitative comparison of E. coli-induced bronchus-associated lymphoid tissue (BALT) in wild type (WT)and Cxcr5 −/− mice. (a) Representative light sheet microscopy (LiSM) micrographs showing the distributions of T and B cell aggregates in the lung on day 18 following E. coli infection. Micrographs were taken from lung lobes stained with anti-CD3 (red) and anti-B220 (blue). (b) Quantitative analysis, using Imaris software, showing the volume of the whole lung lobe occupied by BALT; comparison between WT and Cxcr5 −/− mice. (c) Quantitative analysis of T- and B-cell aggregates in whole lung lobes; comparison between WT and Cxcr5 −/− mice. Data (b, c) was collected from 4 WT and 4 Cxcr5 −/− mice, in which four lung lobes were analyzed per mouse from at least 3 independent experiments. (d) Representative immunofluorescence micrographs taken from lung cryosections of WT and Cxcr5 −/− mice on day 18 following E. coli infection. Cryosections were stained with anti-CD21/35 (green), anti-CD3 (red) and anti-B220 antibodies. (e) Relative frequency of type I, type II and type III lymphoid aggregates on day 18 following E. coli infection of WT and Cxcr5 −/− mice. (f) Sizes of type I, type II and Type III lymphoid aggregates quantified on day 18 following E. coli infection of WT and Cxcr5 −/− mice. Data (e, f) were collected from 7 WT and 4 Cxcr5 −/− mice, in which 4 lung cryosections were analyzed per mouse from at least 3 independent experiments. Bars (b, c, f) represent median values; error bars (e) represent means±s.d.; *P<0.5; ***P<0.001. The color of stars (e) relates to the type of aggregate, as indicated.
BALT in WT mice has been described as cellular aggregates of organized B-cell follicles surrounded by T cells.37 Immunofluorescence micrographs taken from lung sections of Cxcr5 −/− mice showed a different cellular arrangement. A higher number of organized BALT aggregates consisted of T cells in the center, with B cells, which rarely contained FDCs, around them (Figure 6d). The relative frequencies of type I and type III lymphoid aggregates were significantly lower, while the frequency of Type II aggregates was significantly higher in Cxcr5 −/− mice (Figure 6e). Furthermore, all types of lymphoid aggregates were significantly larger in Cxcr5 −/− mice (Figure 6f).
The results from these experiments show that E. coli-induced BALT organization involves the CXCL13/CXCR5 signaling pathways. The organization of BALT in Cxcr5 −/− mice is different from classical BALT, which is defined by B-cell follicles surrounded by T cells. E. coli-induced BALT is characterized by larger individual aggregates containing more T cells that accumulate in the center of BALT, surrounded by FDC-poor B cell zones.
Lack of MyD88-, TRIF-, Cardif- and IFNαR1-mediated signaling does not affect the formation of E. coli-induced BALT
Induction of immune responses requires pattern recognition receptor association with specific pathogen-associated molecular patterns in a process that initiates downstream signaling cascades via adaptor molecules, such as MyD88, TRIF and Cardif.38,39 We reported previously that P. aeruginosa-induced BALT in mice deficient for Myd88 and Trif showed differences regarding the relative frequency and size of lymphoid aggregates compared to WT mice.5 Similarly, we induced E. coli BALT in Myd88 −/− Trif −/−, Myd88 −/− Trif −/− Cardif −/−, and Ifnαr1 −/− mice. We included Ifnαr1 −/− mice because Toll-like receptor (TLR) signaling often results in the production of type I interferons. On day 18 following infection, we analyzed the quality and quantity of induced BALT in the above-mentioned strains, as well as in WT mice. Surprisingly, there were no obvious differences regarding the cellular composition of the strains analyzed, with the exception of Ifnαr1 −/− mice wherein the majority of type I aggregates lacked FDCs (Figure 7a). A similar observation was made regarding the relative frequency of different types of lymphoid aggregates. Here again, the number of type I aggregates in Ifnαr1 −/− mice was significantly reduced (Figure 7b). However, aggregates were larger in both Myd88 −/− Trif −/− and Myd88 −/− Trif −/− Cardif −/− mice compared to WT mice, but similar in Ifnαr1 −/− mice, with the exception of type III aggregates (Figure 7c).
Lack of MyD88, TRIF, Cardif and IFNαR1 signaling does not affect the development of E. coli-induced BALT. (a) Representative immunofluorescence micrographs from lung sections taken from day 18 following E. coli infection in WT, Myd88 −/− Trif −/−, Myd88 −/− Trif −/− Cardif −/− and Ifnαr1 −/− mice. (b) Relative frequency and (c) size of type I, type II and type III lymphoid aggregates on day 18 following E. coli infection. Data (b, c) collected from seven WT, 2 Myd88 −/− Trif −/−, 3 Myd88 −/− Trif −/− Cardif −/− and 3 Ifnαr1 −/− mice, in which four lung sections were analyzed from at least 2 independent experiments. Error bars (b) represent means±s.d.; bars (c) represent median values; *P<0.5; ***P<0.001. The color of stars (b) relates to the type of aggregate, as indicated. Scale bars=100 μm.
These observations show that, in contrast to P. aeruginosa, the induction of BALT by E. coli does not rely on MyD88-, TRIF- or CARDIF-mediated signaling. However, there was a minimal effect of IFNαR1, a receptor that is commonly involved in anti-viral immune responses.
Discussion
This study demonstrates that LiSM can be applied to visualize and quantify BALT within entire lung lobes of mice. We could identify and visualize BALT by staining constituent lymphocytes with antibodies prior to optical clearing and LiSM imaging. Antibody staining and imaging of whole organs have been previously used to study the expression of antigens in embryonic and adult neural organs, to understand disease hallmarks and kidney function.16,40,41 Here, we used a model of spontaneous BALT in Ccr7 −/− mice, as well as BALT induced by pathogens, to determine its volume per lung lobe by computational analysis using Imaris software.
We had previously reported that spontaneous BALT in Ccr7 −/− mice is primarily positioned in specific locations within the lung, that is, close to the main bronchi and surrounding vessels.29 Based on these observations, an immunohistology-based protocol for the qualitative and quantitative characterization of BALT based on the analysis of 4 complete lung sections taken from the central lung harboring the main bronchi has been used to determine the representative quantification of BALT.4,5,29 As a consequence, BALT located at more distal regions was excluded from the analysis. This limitation has been overcome with the protocol reported in this manuscript, and our data show that LiSM can be superbly suited for visualizing and determining the volume of BALT in the entire lung.
Furthermore, we showed that E. coli can induce BALT in mice. Until now, there has been no evidence that intranasal administration of E. coli induces BALT, except where LPS co-application with influenza virus has been used; in that case, it was evident that LPS-induced lung inflammation alone does not lead to the development of BALT, even when administered, repetitively.6 Here, we administered a single dose of heat-inactivated E. coli, lab strain DH5-α, which led to the development of BALT that was characterized by organized B-cell follicles consisting of CXCL13-expressing FDCs and other cells expressing CXCL12. While CXCL12 was present throughout the time course of our analysis, CXCL13 was only present from day 18 onwards, suggesting that both chemokines serve different functions during E. coli-induced BALT formation and maintenance. Unlike in MVA-induced BALT, where the peak time point was reached on day 12 and both CXCL12 and CXCL13 are expressed early,4,5 the peak time point for E. coli-induced BALT was reached at a later time point (day 18). The later emergence of CXCL13 following induction was likely crucial for this long-term maintenance (up to day 30).
The development of primary and secondary lymphoid organs follows a chain of genetically encoded events,42 while the formation of tertiary lymphoid organs, such as induced BALT, is primarily driven by inflammatory or autoimmune-based events. To further understand the role of key chemokines required for the recruitment of lymphocytes during the formation of E. coli-induced BALT, we compared its formation in WT and Cxcr5 −/− mice. Similar to observations in WT mice, E. coli was able to induce development of BALT in Cxcr5 −/− mice, indicating that E. coli BALT induction does not entirely depend on CXCL13/CXCR5-mediated signaling. Related results have been previously reported by us and others for MVA-induced BALT in Cxcr5 −/− mice 5 and influenza-induced BALT in Cxcl13 −/− mice.43 Rangel-Moreno et al 43 reported that BALT in Cxcl13 −/− mice could promote the generation of influenza-specific IgG. In our study, we did not determine whether antibody responses were affected in Cxcr5 −/− mice, but based on similar observations regarding influenza-induced BALT, we speculate that this would not be the case.
However, E. coli-induced BALT in Cxcr5 −/− mice was characterized by T-cell aggregates surrounded by B-cell follicles, in contrast to WT mice where B cells follicles were always located in the center. Further LiSM analysis indicated that the majority of aggregates in Cxcr5 −/− mice were dominated by T cells. In our previous reports concerning secondary lymphoid organs, we observed that the disruption of CXCR5 leads to defects in B-cell migration and follicle organization.22 This effect appears to be the same for tertiary lymphoid organs, based on evidence from E. coli-induced BALT indicated by our results in this study and recent reports using Pneumocystis jirovecii for BALT induction.44
Our results also show that E. coli-induced BALT develops in mice deficient for either two (Myd88 −/− Trif −/−) or three (Myd88 −/− Trif −/− Cardif −/−) adaptor molecules that are associated with signaling pathways initiated by pattern recognition receptors during induction of immune responses to pathogens.38,39 We have previously reported that defects in MyD88 and TRIF (when either one or both molecules were knocked out) impaired the development of P. aeruginosa-induced, but not MVA-induced, BALT.5 Pathogen-associated molecular patterns associated with E. coli, such as LPS, can be recognized by TLR4.45 Despite the fact that MyD88 acts as a general signaling adaptor molecule associated with TLRs,46 a MyD88-independent pathway for TLR4, based on the activation of IRF3 and induction of IFN-inducible genes, has been previously reported.47 Therefore, it can be speculated that the development of E. coli-induced BALT in these mice could involve other different pathways. However, to confirm this hypothesis, additional experiments need to be performed.
IFNαR1 signaling is commonly associated with anti-viral responses.48 However, we observed a slight impairment in E. coli-induced BALT in Ifnαr1 −/− mice. There are reports that show evidence of a protective role played by IFN signaling against E. coli infection, by enhancing the production of TNF-α and IFN-γ.49 It is possible that the slight impairment in E. coli-induced BALT in Ifnαr1 −/− mice could be due to effects on induction of host protective immune response following E. coli administration.
In conclusion, we report a method that allows robust analysis of the architecture of entire lung lobes, including the bronchial and lymphatic vascular tree and structures present in lungs, such as BALT. This could not previously be determined by examining epifluorescence on cryosections. Alone or in combination with immunohistochemistry, this approach will provide additional insights into understanding the development and organization of BALT and may be useful for studying other lung pathologies, such as lung carcinomas. Nevertheless, LiSM has certain limitations. For example, we could not determine the distribution and organization of lymphocytes within BALT and measure their size in our analysis. Furthermore, we could not use LiSM to visualize certain cell populations and chemokines, because they require special staining conditions that, so far, are not compatible with our staining and clearing protocol. In addition, we identified E. coli as a potent inducer of BALT. Considering the enormous volume BALT can reach following pathogen exposure, as demonstrated in this study, it is important to investigate the development of BALT in different induction models and to further delineate its function in initiating adaptive immune responses to control infections of the lung.
References
Sminia T, van der Brugge-Gamelkoorn GJ, Jeurissen SH. Structure and function of bronchus-associated lymphoid tissue (BALT). Crit Rev Immunol 1989; 9: 119–150.
Pabst R, Gehrke I. Is the bronchus-associated lymphoid tissue (BALT) an integral structure of the lung in normal mammals, including humans? Am J Respir Cell Mol Biol 1990; 3: 131–135.
Tschernig T, Pabst R. Bronchus-associated lymphoid tissue (BALT) is not present in the normal adult lung but in different diseases. Pathobiology 2000; 68: 1–8.
Halle S, Dujardin HC, Bakocevic N, Fleige H, Danzer H, Willenzon S et al. Induced bronchus-associated lymphoid tissue serves as a general priming site for T cells and is maintained by dendritic cells. J Exp Med 2009; 206: 2593–2601.
Fleige H, Ravens S, Moschovakis GL, Bölter J, Willenzon S, Sutter G et al. IL-17-induced CXCL12 recruits B cells and induces follicle formation in BALT in the absence of differentiated FDCs. J Exp Med 2014; 211: 643–651.
Rangel-Moreno J, Carragher DM, de la Luz Garcia-Hernandez M, Hwang JY, Kusser K, Hartson L et al. The development of inducible bronchus-associated lymphoid tissue depends on IL-17. Nat Immunol 2011; 12: 639–646.
Moyron-Quiroz J, Rangel-Moreno J, Carragher DM, Randall TD. The function of local lymphoid tissues in pulmonary immune responses. Adv Exp Med Biol 2007; 590: 55–68.
Day TA, Koch M, Nouailles G, Jacobsen M, Kosmiadi GA, Miekley D et al. Secondary lymphoid organs are dispensable for the development of T-cell-mediated immunity during tuberculosis. Eur J Immunol 2010; 40: 1663–1673.
Huisken J. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 2004; 305: 1007–1009.
Keller PJ, Stelzer EH. Quantitative in vivo imaging of entire embryos with digital scanned laser light sheet fluorescence microscopy. Curr Opin Neurobiol 2008; 18: 624–632.
Boas D. A fundamental limitation of linearized algorithms for diffuse optical tomography. Opt Express 1997; 1: 404–413.
Dodt H-U, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 2007; 4: 331–336.
Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc 2012; 7: 1983–1995.
Susaki EA, Tainaka K, Perrin D, Yukinaga H, Kuno A, Ueda HR. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat Protoc 2015; 10: 1709–1727.
Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ et al. Structural and molecular interrogation of intact biological systems. Nature 2013; 497: 332–337.
Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 2014; 159: 896–910.
Förster R, Schubel A, Breitfeld D, Kremmer E, Renner-Müller I, Wolf E et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell 1999; 99: 23–33.
Waibler Z, Anzaghe M, Frenz T, Schwantes A, Pöhlmann C, Ludwig H et al. Vaccinia virus-mediated inhibition of type I interferon responses is a multifactorial process involving the soluble type I interferon receptor B18 and intracellular components. J Virol 2009; 83: 1563–1571.
Spanier J, Lienenklaus S, Paijo J, Kessler A, Borst K, Heindorf S et al. Concomitant TLR/RLH signaling of radioresistant and radiosensitive cells is essential for protection against vesicular stomatitis virus infection. J Immunol 2014; 193: 3045–3054.
Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM et al. Functional role of type I and type II interferons in antiviral defense. Science 1994; 264: 1918–1921.
Bach P, Kamphuis E, Odermatt B, Sutter G, Buchholz CJ, Kalinke U. Vesicular stomatitis virus glycoprotein displaying retrovirus-like particles induce a type I IFN receptor-dependent switch to neutralizing IgG antibodies. J Immunol 2007; 178: 5839–5847.
Förster R, Mattis AE, Kremmer E, Wolf E, Brem G, Lipp M. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell 1996; 87: 1037–1047.
Kremer M, Volz A, Kreijtz JHCM, Fux R, Lehmann MH, Sutter G. Easy and efficient protocols for working with recombinant vaccinia virus MVA. Methods Mol Biol 2012; 890: 59–92.
Worbs T, Mempel TR, Bölter J, von Andrian UH, Förster R. CCR7 ligands stimulate the intranodal motility of T lymphocytes in vivo. J Exp Med 2007; 204: 489–495.
Reynaud EG, Krzic U, Greger K, Stelzer EHK. Light sheet-based fluorescence microscopy: more dimensions, more photons, and less photodamage. HFSP J 2008; 2: 266–275.
Zheng H, Rinaman L. Simplified CLARITY for visualizing immunofluorescence labeling in the develo** rat brain. Brain Struct Funct 2016; 221: 2375–2383.
Richardson DS, Lichtman JW. Clarifying tissue clearing. Cell 2015; 162: 246–257.
Pan C, Cai R, Quacquarelli FP, Ghasemigharagoz A, Lourbopoulos A, Matryba P et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat Methods 2016; 13: 859–867.
Kocks JR, Davalos-Misslitz ACM, Hintzen G, Ohl L, Förster R. Regulatory T cells interfere with the development of bronchus-associated lymphoid tissue. J Exp Med 2007; 204: 723–734.
Glaser K, Speer CP. Toll-like receptor signaling in neonatal sepsis and inflammation: a matter of orchestration and conditioning. Expert Rev Clin Immunol 2013; 9: 1239–1252.
De Lastours V, Malosh RE, Aiello AE, Foxman B. Prevalence of Escherichia coli carriage in the oropharynx of ambulatory children and adults with and without upper respiratory symptoms. Ann Am Thorac Soc 2015; 12: 461–463.
Vincent J-L, Rello J, Marshall J, Silva E, Anzueto A, Martin CD et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA 2009; 302: 2323.
Emori TG, Gaynes RP. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev 1993; 6: 428–442.
Haddad E-B, Birrell M, McCluskie K, Ling A, Webber SE, Foster ML et al. Role of p38 MAP kinase in LPS-induced airway inflammation in the rat. Br J Pharmacol 2001; 132: 1715–1724.
Harmsen AG. Role of alveolar macrophages in lipopolysaccharide-induced neutrophil accumulation. Infect Immun 1988; 56: 1858–1863.
Hajna Z, Borbély É, Kemény Á, Botz B, Kereskai L, Szolcsányi J et al. Hemokinin-1 is an important mediator of endotoxin-induced acute airway inflammation in the mouse. Peptides 2015; 64: 1–7.
Randall TD. Bronchus-associated lymphoid tissue (BALT) structure and function. Adv Immunol 2010; 107: 187–241.
Opitz B, van Laak V, Eitel J, Suttorp N. Innate immune recognition in infectious and noninfectious diseases of the lung. Am J Respir Crit Care Med 2010; 181: 1294–1309.
Takeuchi O, Akira S, Li S, Wang YY, Li Y, Diao F et al. Pattern Recognition Receptors and Inflammation. Cell 2010; 140: 805–820.
Liebmann T, Renier N, Bettayeb K, Greengard P, Tessier-Lavigne M, Flajolet M. Three-dimensional study of Alzheimer’s disease hallmarks using the iDISCO clearing method. Cell Rep 2016; 16: 1138–1152.
Klingberg A, Hasenberg A, Ludwig-Portugall I, Medyukhina A, Männ L, Brenzel A et al. Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. J Am Soc Nephrol 2017; 28: 452–459.
Drayton DL, Liao S, Mounzer RH, Ruddle NH. Lymphoid organ development: from ontogeny to neogenesis. Nat Immunol 2006; 7: 344–353.
Rangel-Moreno J, Moyron-Quiroz JE, Hartson L, Kusser K, Randall TD. Pulmonary expression of CXC chemokine ligand 13, CC chemokine ligand 19, and CC chemokine ligand 21 is essential for local immunity to influenza. Proc Natl Acad Sci USA 2007; 104: 10577–10582.
Eddens T, Elsegeiny W, de la Garcia-Hernadez ML, Castillo P, Trevejo-Nunez G, Serody K et al. Pneumocystis-driven inducible bronchus-associated lymphoid tissue formation requires Th2 and Th17 immunity. Cell Rep 2017; 18: 3078–3090.
Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem 1999; 274: 10689–10692.
Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 2006; 124: 783–801.
Kawai T, Takeuchi O, Fujita T, Inoue J, Mühlradt PF, Sato S et al. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J Immunol 2001; 167: 5887–5894.
Welsh RM, Bahl K, Marshall HD, Urban SL. Type 1 interferons and antiviral CD8 T-Cell responses. PLoS Pathog 2012; 8: e1002352.
Mancuso G, Midiri A, Biondo C, Beninati C, Zummo S, Galbo R et al. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J Immunol 2007; 178: 3126–3133.
Acknowledgements
We thank Stephan Halle, Olga Halle, Annika Reinhardt and **aokun Liu for their help in establishing and troubleshooting the LiSM, as well as Susanne Häußler for constructive comments in this project. This work was supported by a Lichtenberg-stipend to DTM through the University of Veterinary Medicine, Hannover, the German Excellence Initiative grant EXC62 —Rebirth (to RF) and DFG grant SFB 900/B1 (to RF).
Author contributions
DTM performed experiments, analyzed data and wrote the manuscript. HF designed and performed experiments and critically reviewed the manuscript. KL, SW, and JR performed experiments. JS, GS, PV-W and U.K. provided valuable reagents and mice and reviewed the manuscript. RF designed experiments and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
Open Access This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if thematerial is not included under the CreativeCommons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Mzinza, D.T., Fleige, H., Laarmann, K. et al. Application of light sheet microscopy for qualitative and quantitative analysis of bronchus-associated lymphoid tissue in mice. Cell Mol Immunol 15, 875–887 (2018). https://doi.org/10.1038/cmi.2017.150
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cmi.2017.150
- Springer Nature Limited
Keywords
This article is cited by
-
Assessing the combined impact of fatty liver-induced TGF-β1 and LPS-activated macrophages in fibrosis through a novel 3D serial section methodology
Scientific Reports (2024)
-
Optical clearing and testing of lung tissue using inhalation aerosols: prospects for monitoring the action of viral infections
Biophysical Reviews (2022)
-
The chemokine receptor CCR7 is a promising target for rheumatoid arthritis therapy
Cellular & Molecular Immunology (2019)
-
Imaging of Murine Whole Lung Fibrosis by Large Scale 3D Microscopy aided by Tissue Optical Clearing
Scientific Reports (2018)