Abstract
ProNGF signaling through p75NTR has been associated with neurodegenerative disorders. Retinitis pigmentosa (RP) comprises a group of inherited retinal dystrophies that causes progressive photoreceptor cell degeneration and death, at a rate dependent on the genetic mutation. There are more than 300 mutations causing RP, and this is a challenge to therapy. Our study was designed to explore a common mechanism for p75NTR in the progression of RP, and assess its potential value as a therapeutic target. The proNGF/p75NTR system is present in the dystrophic retina of the rd10 RP mouse model. Compared with wild-type (WT) retina, the levels of unprocessed proNGF were increased in the rd10 retina at early degenerative stages, before the peak of photoreceptor cell death. Conversely, processed NGF levels were similar in rd10 and WT retinas. ProNGF remained elevated throughout the period of photoreceptor cell loss, correlating with increased expression of α2-macroglobulin, an inhibitor of proNGF processing. The neuroprotective effect of blocking p75NTR was assessed in organotypic retinal cultures from rd10 and RhoP mouse models. Retinal explants treated with p75NTR antagonists showed significantly reduced photoreceptor cell death, as determined by the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay and by preservation of the thickness of the outer nuclear layer (ONL), where photoreceptor nuclei are located. This effect was accompanied by decreased retinal-reactive gliosis and reduced TNFα secretion. Use of p75NTR antagonist THX-B (1,3-diisopropyl-1-[2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-purin-7-yl)-acetyl]-urea) in vivo in the rd10 and RhoP mouse models, by a single intravitreal or subconjunctival injection, afforded neuroprotection to photoreceptor cells, with preservation of the ONL. This study demonstrates a role of the p75NTR/proNGF axis in the progression of RP, and validates these proteins as therapeutic targets in two different RP models, suggesting utility irrespective of etiology.
Similar content being viewed by others
Main
Retinitis pigmentosa (RP) refers to a group of inherited retinal dystrophies that are clinically similar despite arising from a large set of genetic mutations (http://www.sph.uth.tmc.edu/Retnet/disease.htm). These mutations usually trigger photoreceptor cell degeneration and death, leading to visual function decline and, eventually, blindness.1 While the time of onset and the rate of neurodegeneration are specified by the mutation, most, if not all, forms of RP share molecular and cellular mechanisms that include inflammation, microglial activation and reactive gliosis. These features are shared with other retinal diseases without a pure genetic origin, such as glaucoma, diabetic retinopathy and age-related macular degeneration.2, 3
Various therapeutic strategies for RP, including gene, cell and regenerative therapies, as well as pharmacological treatments, are gradually progressing from the animal models to clinical trials4, 5, 6 and http://clinicaltrials.gov/ct2/results?term=retinitis+pigmentosa. However, there is not yet any approved treatment for the neurodegenerative component of retinal diseases. The genetic complexity in the etiology of RP, comprising more than 300 described mutations in over 50 different genes, calls for the development of treatments targeting common mechanisms independently of the causative mutation. This would entail the detailed characterization of the processes leading to retinal deterioration, as a strategy to discover novel therapeutic targets.
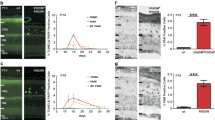
As a monogenic genetic disease of high penetrance, a variety of animal genetic models recapitulate the signs and symptoms of human RP.4 A missense mutation in the Pde6b gene causes blindness in the rd10 mouse model of autosomal recessive RP.7 The RhoP mouse model of autosomal dominant RP carries the mutant human rhodopsin Pro347Ser transgene.8 The course of the disease in these models recapitulate human progression and allows for effective experimental interventions.9, 10, 11, 12, 13, 33 as well as in models of retinal dystrophies.34 In the rd10 retina, microglia is highly activated and mobilized (Figures 2d–f), and may be the source of proNGF.
ProNGF is efficiently processed in the uninjured adult nervous system and very low levels of the proprotein form are detected under physiological conditions. The intracellular machinery that ensures the efficient conversion of proNGF into mature NGF is not well characterized. Retinal degeneration in the rd10 mouse was accompanied by an increase of α2M, selectively located to the photoreceptor segment (Figures 2b and c and Supplementary Figure 2) and presumptively expressed by Müller glia cells,35 whose role in retinal neurodegeneration has been recently characterized in detail.22, 24 α2M binds to proNGF conferring resistance to processing and increased activity on p75NTR. The marked increase in the α2M in the rd10 retina may account, at least in part, for the accumulation of proNGF. On the other hand, α2M binds to NGF and decreases its activity on the Trk-A receptor.
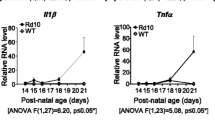
A hallmark of most forms of RP, despite their genetic origin, is the primary death of rod photoreceptors, in which the mutated gene exerts its function, followed by a secondary loss of cone photoreceptors.5 Photoreceptor degeneration is accompanied and followed by whole retinal remodeling, including retinal pigmented epithelium disorganization, inner neuronal connectivity alterations and vascular disorganization and regression. Concomitant to these alterations an inflammatory process takes place as shown by microglial recruitment, reactive gliosis of Müller glial cells and astrocytes, and a burst of proinflammatory cytokine expression.3 The hierarchies, respective roles and molecular action of microglia, Müller cells and astrocytes during rd10 retinal degeneration remain to be elucidated. Interfering with one or several of these common players may provide a general therapy, despite the broad mutation spectrum of RP. Supporting this approach, suppression of microglial activation by minocycline treatment protected retinal structure and visual function in the rd10 mouse.14 Further, TNFα inhibition by Adalimumab in the rd10 mouse reduces photoreceptor cell death.16
In the present study, we have extended to RP the therapeutic potential of interfering with the proNGF/p75NTR system described for other retinal dystrophies.20, 21, 31
Materials and methods
Animal procedures
The rd10 mouse model of retinal degeneration is a homozygous recessive mutant for phosphodiesterase 6b (Pde6brd10/rd10) on a C57BL/6 J background. The RhoP mouse model of retinal degeneration is a hemizygous dominant mutant that carries the mutant human rhodopsin Pro347Ser transgene on a C57BL/6 J background. WT mice of the same background were used as control. All animals were housed and handled in accordance with the ARVO statement for the Use of Animals in Ophthalmic and Vision Research, and European Union and Canadian guidelines. Mice were bred in the CIB and Lady Davis Institute core facilities.
Intravitreal injections
Rd10 and RhoP mice at P17 were anesthetized with isoflurane. Intravitreal injection was performed under an ophthalmoscope to visualize the retinal fundus. Using a Hamilton syringe (Hamilton AG, Bonaduz, Switzerland; Model 75 RN SYR) with a 33-gauge removable needle, the right eye was injected with 2 μl of the p75NTRantagonist, THX-B (2 μg/μl in 10% (v/v) DMSO in PBS), whereas the left eyes received 2 μl of the vehicle. Animals were killed 5 days after injection, and eyes were processed for cryosectioning as described below. Both male and female mice were used for this study and at least three animals per group were used.
Subconjunctival injection
RhoP at P18 were anesthetized with isoflurane. The conjunctiva was gently pulled from the sclera using tweezers. The conjunctiva of the right eye was injected with 30 μl containing a total of 15 μg THX-B (0.5 μg/μl in 10% (v/v) DMSO in PBS). Half of the total volume of THX-B was delivered into the superior subconjunctival space and the other half into the nasal subconjunctival region using a microsyringe with a 33 G needle. The left eyes received 30 μl of the vehicle. Animals were killed 4 days after injection and processed as above.
p75NTR antagonists synthesis
THX-B and the analogs have been described20 (US20100392647P patent by H Nedev and HU Saragovi). All compounds were purified by HPLC to >99%. The empirical formula of THX-B (F.W. 365) was confirmed by LC/MS, and the 1H NMR spectra (300 MHz) of compounds are consistent with the expected structures.
Retina explant cultures
P20 and P22 rd10 and P18 RhoP mice were killed, and their eyes were enucleated. Retinas were dissected and cultured free floating in M24 multiwell plates for 24 h in 1 ml DMEM/F12 medium containing N2 supplement, except insulin, and in the presence of the indicated p75NTR antagonist THX-A, THX-B, THX-C, LM11A-31 (Tocris, Avonmouth, Bristol, UK), or vehicle. A dose range of 0.2–40 μM drug concentrations were tested (only 20 μM shown). Retinas were subsequently fixed in 4% (wt/vol) paraformaldehyde in phosphate buffer 0.1M, pH 7.4, for 1 h at RT and processed for detection of cell death and immunostaining.
Cell death visualization and counting
Photoreceptor cell death was visualized by DNA fragmentation using the TUNEL assay (DeadEnd Fluorometric TUNEL System; Promega, Madison, WI, USA). Fixed whole-mount retinas were permeated with 2% (wt/vol) Triton X-100 (Merck, Darmstadt, Germany) in PBS for 2 h at RT, followed by incubation in 20 μg/ml proteinase K (Merck) for 15 min at 37 °C and, subsequently, processed for TUNEL staining according to the manufacturer's instructions. After TUNEL labeling, the nuclei were counterstained with 0.05% (wt/vol) propidium iodide (Merck) in PBS, and the retinas were flat mounted in Fluoromount-G (Southern Biotechnology, Birmingham, AL, USA), and analyzed with a laser confocal microscope (TCS SP2; Leica Microsystems, Wetzlar, Germany). Serial optical sections were acquired with the × 63 objective every 1 μm, covering the whole ONL thickness in four central fields around the optic nerve. Direct counting of TUNEL-positive cells was carried out using the FIJI open-source Software.36
Immunolabeling of whole-mount retinas
Fixed whole-mount retinas were permeated with 2% (wt/vol) Triton X-100 in PBS, blocked in BGT (2.5% (wt/vol) BSA, 100 mM glycine, 0.25% (wt/vol) Triton X-100 in PBS) and incubated overnight at 4 °C, with the indicated primary antibodies or 2 h at RT with a biotinylated probe, after washing with TBS. Retinas were then washed with PBS and incubated for 2 h at RT with the secondary antibodies or streptavidin. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific, Waltham, MA USA), the retinas were mounted in Fluoromount-G and analyzed with a laser confocal microscope (TCS SP5 and TCS SP2; Leica Microsystems). Antibodies are listed below.
For determining microglia density, serial optical sections were acquired with a × 40 objective every 2.5 μm through the whole retina in four central fields around the optic nerve. To measure the percentage of the area occupied by GFAP staining, serial optical sections were acquired with a × 40 objective every 2 μm, covering the whole ganglion cell layer in 12 fields (four central, four medial and four peripheral) around the optic nerve. Confocal maxima of the ganglion cell layer Z sections were then converted into a black and white image and analyzed with the FIJI Software to calculate the percentage of total area filled with GFAP.
Immunostaining of retinal sections
Animals were killed, their eyes were enucleated, fixed for 1 h in 4% (wt/vol) parafolmaldehyde in phosphate buffer 0.1 M, pH 7.4, and then cryoprotected by incubation in increasing concentrations of sucrose (15–50% in PBS). The eyes were then embedded in Tissue-Tek (Sakura, Leiden, The Netherlands), and frozen on dry ice. Cryostat sections (12 μm) were mounted on Superfrost slides (Thermo Fisher Scientific) and dried at room temperature. For p75NTR immunostaining, retinal sections were rehydrated in PBS, incubated with NaBH4 5 mg/ml in PBS for 5 min, followed by 0.1% (wt/vol) Triton X-100 and 5% (v/v) normal goat serum in PBS for 1 h. For immunostaining with the rest of the antibodies, retinal sections were permeated for 1 h with 0.2% (wt/vol) Triton X-100, and blocked with BGT for another hour. Following the incubation with the primary antibody overnight at 4 °C, the sections were washed with PBS and incubated for 1 h with secondary antibodies and DAPI. Then, sections were washed with PBS and coverslipped with Fluoromont-G. Antibodies are listed below.
To quantify the retinal structure using techniques unbiased to retinal location (i.e. the central regions of the retina are thicker than the peripheral regions), we compared the thickness of the ONL (containing primarily photoreceptors) and the corresponding INL (containing bipolar, horizontal and amacrine neuron as well as Müller glia cell bodies). Both ONL and INL were measured, and the ONL/INL ratios were calculated in treated eyes versus vehicle-injected contralateral eyes. Five sections per retina were analyzed. For each section, one photograph was taken for each of four defined retinal zones. In each photo three measurements of the ONL and INL thickness were performed in triplicate to obtain an average value per retinal zone per section. The measurements were carried out with the FIJI Software (using the ‘freehand line’ and ‘measure’ tools). Alternatively, the number of nucleus rows or the nuclear density in a determined retinal area were quantified in the same way to corroborate the neuroprotective effect and the accuracy of the ONL/INL ratio determination.

ProNGF, NGF and TNFα levels determination
ProNGF and NGF levels were measured in retinal extracts from the same animals by ELISA assay (MyBioSource Inc., San Diego, CA, USA). For proNGF determination, four retinas per age were homogenized in PBS at 100 mg wet weight per ml and stored overnight at −20 °C. After two freeze–thaw cycles, the homogenates were centrifuged for 5000xg at 4 °C for 5 min. The supernatant was removed and assayed immediately following the manufacturer’s instructions. For NGF determination, the four contralateral retinas per age of the same animal used for proNGF determination were homogenized in PBS at 100 mg wet weight per ml and centrifuged for 1500xgfor 15 min. The supernatant was removed and assayed immediately. TNFα concentration was measured in culture supernatant from retinal explants by ELISA assay (BioLegend, San Diego, CA, USA) following the manufacturer’s instructions.
RNA isolation and quantitative PCR
Total RNA from individual retinas was extracted using the TRIzol reagent, and 2.5 μg of RNA were typically reverse transcribed (RT) using the Superscript III Kit and random primers (all from Thermo Fisher Scientific). Quantitative PCR (qPCR) was performed with the ABI Prism 7900HT Sequence Detection System using TaqMan Universal PCR Master Mix, No-AmpErase UNG and the Taqman assays (listed below) for detection (all from Thermo Fisher).

Statistical analysis
Data size was estimated in accordance with previous literature, following the Reduction principle for animal research. At least three animals were analyzed per experimental point. Data points in graphs represent individual mice, and bars in all panels represent the mean and the standard error of the mean (S.E.M.).
Data were checked for normality using both D’Agostino–Pearson omnibus and Kolmogorov–Smirnov normality tests sequentially. Data were considered to fit a normal distribution only if they passed simultaneously both tests. For normal data, Fisher’s test was used to determine whether the variance of the samples analyzed was comparable (homoscedasticity). Normal data were compared using a two-way ANOVA with Bonferroni's multiple comparison test or an unpaired Student’s T-test, applying Welch’s correction in cases of non-homoscedasticity. In cases of non-normal samples, populations were compared using the Mann–Whitney nonparametric U-test. Outliers were detected by Grubbs’ outlier test, and excluded from further analysis. If previous literature allowed us to make a prediction about the result of the experiment, then one-sided test were applied. Otherwise, tests were two sided. All analyses were performed at a fixed 95% confidence interval, using the GraphPad Prism version 5.01 for Windows (GraphPad Software, San Diego, CA, USA; http://www.graphpad.com). Statistically significant differences are indicated as follows: *P<0.05; **P<0.01; ***P<0.001.
References
Hartong DT, Berson EL, Dryja TP . Retinitis pigmentosa. Lancet 2006; 368: 1795–1809.
Chinskey ND, Besirli CG, Zacks DN . Retinal cell death and current strategies in retinal neuroprotection. Curr Opin Ophthalmol 2014; 25: 228–233.
Cuenca N, Fernandez-Sanchez L, Campello L, Maneu V, De la Villa P, Lax P et al. Cellular responses following retinal injuries and therapeutic approaches for neurodegenerative diseases. Progr Retin Eye Res 2014; 43: 17–75.
Rivas MA, Vecino E . Animal models and different therapies for treatment of retinitis pigmentosa. Histol Histopathol 2009; 24: 1295–1322.
Bramall AN, Wright AF, Jacobson SG, McInnes RR . The genomic, biochemical, and cellular responses of the retina in inherited photoreceptor degenerations and prospects for the treatment of these disorders. Annu Rev Neurosci 2010; 33: 441–472.
Sacchetti M, Mantelli F, Merlo D, Lambiase A . Systematic review of randomized clinical trials on safety and efficacy of pharmacological and nonpharmacological treatments for retinitis pigmentosa. J Ophthalmol 2015; 2015: 737053.
Chang B, Hawes NL, Pardue MT, German AM, Hurd RE, Davisson MT et al. Two mouse retinal degenerations caused by missense mutations in the beta-subunit of rod cGMP phosphodiesterase gene. Vision Res 2007; 47: 624–633.
Li T, Snyder WK, Olsson JE, Dryja TP . Transgenic mice carrying the dominant rhodopsin mutation P347S: evidence for defective vectorial transport of rhodopsin to the outer segments. Proc Natl Acad Sci USA 1996; 93: 14176–14181.
Corrochano S, Barhoum R, Boya P, Arroba AI, Rodriguez-Muela N, Gomez-Vicente V et al. Attenuation of vision loss and delay in apoptosis of photoreceptors induced by proinsulin in a mouse model of retinitis pigmentosa. Invest Ophthalmol Vis Sci 2008; 49: 4188–4194.
Chadderton N, Millington-Ward S, Palfi A, O'Reilly M, Tuohy G, Humphries MM et al. Improved retinal function in a mouse model of dominant retinitis pigmentosa following AAV-delivered gene therapy. Mol Ther 2009; 17: 593–599.
Bramall AN, Szego MJ, Pacione LR, Chang I, Diez E, D'Orleans-Juste P et al. Endothelin-2-mediated protection of mutant photoreceptors in inherited photoreceptor degeneration. PLoS ONE 2013; 8: e58023.
Barone I, Novelli E, Strettoi E . Long-term preservation of cone photoreceptors and visual acuity in rd10 mutant mice exposed to continuous environmental enrichment. Mol Vis 2014; 20: 1545–1556.
Jiang K, Wright KL, Zhu P, Szego MJ, Bramall AN, Hauswirth WW et al. STAT3 promotes survival of mutant photoreceptors in inherited photoreceptor degeneration models. Proc Natl Acad Sci USA 2014; 111: E5716–E5723.
Peng B, **ao J, Wang K, So KF, Tipoe GL, Lin B . Suppression of microglial activation is neuroprotective in a mouse model of human retinitis pigmentosa. J Neurosci 2014; 34: 8139–8150.
Rodriguez-Muela N, Hernandez-Pinto AM, Serrano-Puebla A, Garcia-Ledo L, Latorre SH, de la Rosa EJ et al. Lysosomal membrane permeabilization and autophagy blockade contribute to photoreceptor cell death in a mouse model of retinitis pigmentosa. Cell Death Differ 2015; 22: 476–487.
Martinez-Fernandez de la Camara C, Hernandez-Pinto AM, Olivares-Gonzalez L, Cuevas-Martin C, Sanchez-Arago M, Hervas D et al. Adalimumab reduces photoreceptor cell death in a mouse model of retinal degeneration. Sci Rep 2015; 5: 11764.
Ibanez CF, Simi A . p75 neurotrophin receptor signaling in nervous system injury and degeneration: paradox and opportunity. Trends Neurosci 2012; 35: 431–440.
Hempstead BL . Deciphering proneurotrophin actions. Handbook Exp Pharmacol 2014; 220: 17–32.
Lebrun-Julien F, Morquette B, Douillette A, Saragovi HU, Di Polo A . Inhibition of p75(NTR) in glia potentiates TrkA-mediated survival of injured retinal ganglion cells. Mol Cell Neurosci 2009; 40: 410–420.
Bai Y, Dergham P, Nedev H, Xu J, Galan A, Rivera JC et al. Chronic and acute models of retinal neurodegeneration TrkA activity are neuroprotective whereas p75NTR activity is neurotoxic through a paracrine mechanism. J Biol Chem 2010; 285: 39392–39400.
Mysona BA, Al-Gayyar MM, Matragoon S, Abdelsaid MA, El-Azab MF, Saragovi HU et al. Modulation of p75(NTR) prevents diabetes- and proNGF-induced retinal inflammation and blood-retina barrier breakdown in mice and rats. Diabetologia 2013; 56: 2329–2339.
Barcelona PF, Saragovi HU . A pro-nerve growth factor (proNGF) and NGF binding protein, alpha2-macroglobulin, differentially regulates p75 and TRKa receptors and is relevant to neurodegeneration ex vivo and in vivo. Mol Cell Biol 2015; 35: 3396–3408.
Barcelona PF, Sitaras N, Galan A, Esquiva G, Jmaeff S, Jian Y et al. p75NTR and its ligand proNGF activate paracrine mechanisms etiological to the vascular, inflammatory, and neurodegenerative pathologies of diabetic retinopathy. J Neurosci 2016; 36: 8826–8841.
Fabrizi C, Businaro R, Lauro GM, Fumagalli L . Role of alpha2-macroglobulin in regulating amyloid beta-protein neurotoxicity: protective or detrimental factor? J Neurochem 2001; 78: 406–412.
Arroba AI, Alvarez-Lindo N, van Rooijen N, de la Rosa EJ . Microglia-mediated IGF-I neuroprotection in the rd10 mouse model of retinitis pigmentosa. Invest Ophthalmol Vis Sci 2011; 52: 9124–9130.
Yang T, Knowles JK, Lu Q, Zhang H, Arancio O, Moore LA et al. Small molecule, non-peptide p75 ligands inhibit Abeta-induced neurodegeneration and synaptic impairment. PLoS ONE 2008; 3: e3604.
Galan A, Barcelona PF, Nedev H, Sarunic MV, Jian Y, Saragovi HU . Subconjunctival delivery of p75NTR antagonists reduces the inflammatory, vascular, and neurodegenerative pathologies of diabetic retinopathy. Invest Ophthalmol Vis Sci 2017; 58: 2852–2862.
Nakamura K, Harada C, Okumura A, Namekata K, Mitamura Y, Yoshida K et al. Effect of p75NTR on the regulation of photoreceptor apoptosis in the rd mouse. Mol Vis 2005; 11: 1229–1235.
Al-Shawi R, Hafner A, Chun S, Raza S, Crutcher K, Thrasivoulou C et al. ProNGF, sortilin, and age-related neurodegeneration. Ann NY Acad Sci 2007; 1119: 208–215.
Liepinsh E, Ilag LL, Otting G, Ibanez CF . NMR structure of the death domain of the p75 neurotrophin receptor. EMBO J 1997; 16: 4999–5005.
Lebrun-Julien F, Bertrand MJ, De Backer O, Stellwagen D, Morales CR, Di Polo A et al. ProNGF induces TNFalpha-dependent death of retinal ganglion cells through a p75NTR non-cell-autonomous signaling pathway. Proc Natl Acad Sci USA 2010; 107: 3817–3822.
Frade JM, Rodriguez-Tebar A, Barde YA . Induction of cell death by endogenous nerve growth factor through its p75 receptor. Nature 1996; 383: 166–168.
Frade JM, Barde YA . Microglia-derived nerve growth factor causes cell death in the develo** retina. Neuron 1998; 20: 35–41.
Srinivasan B, Roque CH, Hempstead BL, Al-Ubaidi MR, Roque RS . Microglia-derived pronerve growth factor promotes photoreceptor cell death via p75 neurotrophin receptor. J Biol Chem 2004; 279: 41839–41845.
Shi Z, Rudzinski M, Meerovitch K, Lebrun-Julien F, Birman E, Di Polo A et al. Alpha2-macroglobulin is a mediator of retinal ganglion cell death in glaucoma. J Biol Chem 2008; 283: 29156–29165.
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–682D.
Acknowledgements
This work was supported by the Ministerio de Economía y Competitividad, Spain (Grants SAF2013-41059- R and SAF2016-75681- R to EJdlR), Ministerio de Educación, Cultura y Deporte, Spain (Travel grant to EJdlR), and by CIHR grants (Pharmacology and Proof of Principle) and by the Foundation Fighting Blindness to HUS. MPC has a fellowship from the Ministerio de Economía y Competitividad, Spain. We thank laboratory members for continuous encouragement and ideas, and Cayetana Murillo, José Luis Martínez and the staff of the CIB animal house and microscopy facilities for technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
McGill University has filed patents for the compounds used in this project (HUS inventor).
Additional information
Edited by A Yaron
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Platón-Corchado, M., Barcelona, P., Jmaeff, S. et al. p75NTR antagonists attenuate photoreceptor cell loss in murine models of retinitis pigmentosa. Cell Death Dis 8, e2922 (2017). https://doi.org/10.1038/cddis.2017.306
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2017.306
- Springer Nature Limited
This article is cited by
-
Modulation of GSK-3 provides cellular and functional neuroprotection in the rd10 mouse model of retinitis pigmentosa
Molecular Neurodegeneration (2018)
-
RETRACTED ARTICLE: Non-coding RNAome of RPE cells under oxidative stress suggests unknown regulative aspects of Retinitis pigmentosa etiopathogenesis
Scientific Reports (2018)
-
In retinitis pigmentosa TrkC.T1-dependent vectorial Erk activity upregulates glial TNF-α, causing selective neuronal death
Cell Death & Disease (2017)