Abstract
Death effector domains (DEDs) are protein–protein interaction domains initially identified in proteins such as FADD, FLIP and caspase-8 involved in regulating apoptosis. Subsequently, these proteins have been shown to have important roles in regulating other forms of cell death, including necroptosis, and in regulating other important cellular processes, including autophagy and inflammation. Moreover, these proteins also have prominent roles in innate and adaptive immunity and during embryonic development. In this article, we review the various roles of DED-containing proteins and discuss recent developments in our understanding of DED complex formation and regulation. We also briefly discuss opportunities to therapeutically target DED complex formation in diseases such as cancer.
Similar content being viewed by others
Facts
-
FADD, FLIP, procaspase-8 and procaspase-10 all contain death effector domains (DEDs).
-
The DED is a conserved protein sub-domain that mediates important protein–protein interactions.
-
DED-containing proteins form a variety of complexes that regulate key cellular processes, most notably apoptosis, necroptosis and autophagy.
-
Recent reports also highlight the critical role of DED proteins in other key processes linked to development and inflammation.
Open Questions
-
Does caspase-10 (absent in mice) have overlap** functions with caspase-8, or is it functionally distinct?
-
Under what physiologically relevant conditions does necroptosis occur rather than apoptosis?
-
In which cellular contexts are FADD, FLIP, (pro)caspase-8 and (pro)caspase-10 critical for regulating autophagy?
-
What are the best ways of targeting DED-containing proteins to therapeutically activate cell death (e.g., in cancers) or prevent cell death (e.g., in neurodegenerative diseases)?
-
Are DED-containing proteins potential therapeutic targets for inflammatory diseases?
Cell death is critical for maintaining homeostasis in multicellular organisms; too much can result in pathologies such as neurodegeneration, whereas too little can lead to the accumulation of malignant cancerous cells. Cell death can be either active, where the cell participates in its own destruction or passive, for example, when a cell undergoes irreparable physical damage.1 The most biochemically well-characterised form of cell death is apoptosis, an active process in which cysteine-dependent aspartate-directed proteases (caspases) are activated in response to extracellular stimuli or internal damage culminating in a form of cell death defined by distinct molecular events and characteristic changes in the morphology of the dying cell. Recently, a number of actively regulated non-apoptotic mechanisms of cell death have emerged, including necroptosis, pyroptosis and ferroptosis, which have been comprehensively reviewed elsewhere.2, 3 Here, we focus on those mechanisms of cell death arising following stimulation of death receptors, broadly termed the ‘extrinsic pathway’. For authoritative reviews on mitochondrial-mediated ‘intrinsic’ cell death, we direct the Reader elsewhere.4, 5 Central to receptor-mediated cell death pathways are proteins containing ‘death–fold superfamily’ interaction motifs such as the death domain (DD), caspase activation and recruitment domain (CARD), pyrin domain and the death effector domain (DED).
The DED-containing proteins, which are key decision makers in determining the life and death of cells, are the primary focus of this review. We will first introduce the main members of the DED protein family and discuss advances in the understanding of the assembly and stoichiometry of death receptor complexes. We will then summarise the recent literature surrounding the regulation of these complexes and consider the role of these proteins in disease.
The DED Proteins
The death-fold motif is characterised by its globular structure containing six amphipathic α-helices that run anti-parallel in α-helical bundles.6 When folded, a conserved hydrophobic core forms, although differences in helical length and residue distribution give rise to significant variations between the different sub-families.6, 7, 8, 9 The DED death-fold sub-family consists of procaspases-8 and -10, FLIP, FADD, DEDD, DEDD2, and PEA-15 (Figure 1). FADD, DEDD, DEDD2 and PEA-15 contain a single DED, whereas FLIP, procaspase-8 and procaspase-10 each have tandem DEDs. Procaspases-8 and -10 have catalytically active domains in the regions C-terminal to their tandem DEDs; whereas the long splice form of FLIP, FLIPL, has a pseudo-caspase domain C- terminal to its tandem DEDs, in which the cysteine residue critical for enzymatic activity is absent. Shorter splice forms of FLIP (FLIP short, FLIPS, and FLIP Raji, FLIPR) arising through alternative mRNA splicing lack the pseudo-caspase domain, but contain the tandem DEDs.10
Schematic representations of the structures of DED proteins (ribbon format with transparent solvent-accessible surface area) along with their complete linear domain organisation. Protein structures: (a) Homology model of human FLIP DEDs48 (b) NMR solution structure of FADD DED (PDB ID: 2GF5) 7 (c) NMR solution structure of PEA-15 DED (PDB ID: 2LS7)191 (d) Homology model of procaspase-8 DEDs48 (e) Homology model of procaspase-10 DEDs generated using the I-TASSER web server192
Procaspase-8 is a highly conserved protease, displaying ~20% sequence similarity to its Caenorhabditis elegans homolog CED-3.11 Eight splice forms of procaspase-8 have been identified at the mRNA level, although only two of these, procaspases-8A and 8B are expressed as functional proteases.12 Additionally, a long splice form (procaspase-8L), which contains a 136-bp insert between exons 8 and 9 encoding an early stop codon, contains both DEDs but lacks a functional catalytic domain; it is found in undifferentiated cells and neoplasms and has been reported to act in a dominant negative manner to inhibit apoptosis.13, 14, 15
Procaspase-10 is also expressed as multiple splice forms: procaspases-10A, B, D and G. All contain tandem DEDs and proteolytic domains except G, which is truncated and only consists of the DEDs.16, 17, 18 In vitro studies have shown that procaspase-10 is activated by induced proximity in a manner similar to procaspase-8.19 Despite the similarity between procaspase-10 and procaspase-8, whether procaspase-10 can initiate death receptor-mediated apoptosis in the absence of procaspase-8 remains controversial,20, 21, 22 although they share common substrates, notably BID and RIPK1.23, 24 However, no ortholog of the gene encoding procaspase-10 (Casp10) is present in the mouse genome,25 suggesting that procaspase-10 is not required for activation of the extrinsic apoptotic pathway. Despite this, expression of both procaspase-8 and procaspase-10 is frequently downregulated in cancer.26, 27, 28 Interestingly, the genes encoding procaspase-8 (Casp8), procaspase-10 and FLIP (CFLAR) are present at the same loci (2q33-q34) and clearly evolved via gene duplication events. Evolutionary studies have identified the predecessors of Casp8, Casp10 and CFLAR in fish.29
FADD is an adaptor protein containing a DD which allows it to associate with the DDs of TRAIL- R1, TRAIL-R2, CD95 and TRADD, and a DED, which enables it to recruit other DED-containing proteins, namely procaspase-8, procaspase-10 or FLIP. As with most other components of the extracellular apoptosis signalling pathways, FADD is highly evolutionarily conserved.30 As a protein linking death receptors to death initiators, it is not only a key player in cell death, but also has reported roles in non-apoptotic processes. For example, FADD has been identified in the nucleus and has been postulated to have functions in regulating cell cycle progression,31, 32 NF-κB activity,33 autophagy,34 cytokine signalling35, 36 and T-cell activation.37 Indeed, roles beyond core apoptosis signalling have also been identified for FLIP, procaspase-8 and procaspase-10, and frequently these roles involve complex interactions between these proteins and FADD.
Assembly of the Death-inducing Signalling Complex (DISC)
The death receptors TRAIL-R1 (DR4), TRAIL-R2 (DR5) and CD95 (Fas) are specialised members of the TNF receptor superfamily and are key mediators of apoptosis triggered by ligands expressed by cells of the immune system, namely TRAIL (TNF-related apoptosis-inducing ligand), which activates TRAIL-R1 and TRAIL-R2, and CD95L (FasL), which activates CD95. Following extracellular ligand binding, pre-associated TRAIL-R1, TRAIL-R2 and CD95 trimers, cluster through interactions between their intracellular DDs.38, 39 The DD of FADD can then interact with the cytoplasmic DDs of the death receptors, after which its DED becomes available for protein–protein interactions with other DED proteins, thereby creating a platform for assembly of the DISC.
Death domain interactions
The interactions between the CD95 and FADD DDs have been described (Figure 2). Scott et al.40 reported a 2.7 Å resolution co-crystal structure, which suggests that CD95 and FADD bind in dimeric units, that is 2 × FADD-DD interacting with 2 × CD95-DD. These units are then proposed to further associate into tetrameric structures (4 × FADD-DD to 4 × CD95-DD), although in vivo, the dimeric form is favoured. They report that, following CD95 receptor activation, CD95 undergoes a conformational change, exposing its hydrophobic core and revealing a multitude of interaction surfaces capable of binding the DD of FADD. This represents a possible safety mechanism whereby the apoptotic cascade only proceeds when sufficient CD95 DDs cluster. However, the FADD interaction sites in the CD95 DD predicted by this model do not correlate with mutations observed in patients with autoimmune lymphoproliferative syndrome, a disease defined by mutations in CD95 which prevent DISC formation.41 In a different study, Wang et al.42 used electron microscopy to visualise CD95 DD–FADD DD interactions and observed that they bore a striking resemblance to the PIDD DD–RAIDD DD complex, being principally composed of 5 × CD95 DDs and 5 × FADD DDs layered together. This stoichiometry is in agreement with data by Esposito et al.,43 who also reported a ratio of 5 × CD95:5 × FADD together with some 6 × CD95:5 × FADD and 7 × CD95:5 × FADD ratios, but not the 4 × CD95:4 × FADD suggested by Scott et al. Disparities between these models could be explained by the different conditions used for protein crystallisation; however, the models proposed by Wang et al. and Esposito et al. are supported by the fact that they account for mutations frequently seen in autoimmune lymphoproliferative syndrome. Most disease-causing mutations present in autoimmune lymphoproliferative syndrome patients reside in the DD of CD95,41 resulting in an inability to bind FADD and form a DISC. Map** these mutated residues onto the structure proposed by Wang et al. reveals that they reside on the exposed surface of the DD and would be likely to prevent the binding of FADD.42
Crystal structure of the human CD95–FADD DD heterodimer. CD95 DD (blue ribbon) binds to FADD DD (green ribbon) via several key interacting residues (highlighted as grey sticks) (PDB ID: 3EZQ).40 Hydrogen bonds are depicted as black dotted lines. These heterodimers oligomerise into higher order structures
DED interactions
In addition to death receptor:FADD DD interactions, FADD has been reported to self-associate through its DED, which stabilises its association with the death receptor. Sandu et al.44 identified a ‘hydrophobic patch’ (F25, L28 and K33) as the critical surface for FADD-FADD interactions and not an RxDL motif as had previously been reported. Such interactions between FADD molecules generates higher order complexes of FADD and death receptors that may be the basis of the SPOTS (signaling protein oligomerisation transduction structures), which have been reported to form soon after CD95 receptor activation.45 The RxDL motif is found in both DEDs of viral FLIP MC159 and is critical for its ability to be recruited to the DISC and inhibit apoptosis.9, 46, 47 However, this appears not to be the case for murine FLIP, for which the hydrophobic patch was instead found to be indispensable for DISC recruitment and apoptosis inhibition.47 MC159 was also reported to interact with the RxDL motif of FADD, blocking FADD self-association and preventing the formation of a competent caspase-recruiting platform.8 The role of the RxDL motif may differ for cellular and viral forms of FLIP. Our recent data suggest that this motif is important for human FLIP’s anti-apoptotic function; however, not because of its direct involvement in mediating inter- and intra-molecular interactions, but rather because it controls the spatial orientation of the hydrophobic patch defined by the α2 and α5 helices of FLIP’s DEDs (Figure 3); our data and those of others indicate that it is this hydrophobic patch that mediates intra-molecular interactions between FLIP’s tandem DEDs and inter-molecular interactions between FLIP and FADD and procaspase-8 (Figure 4).48
Conserved features of the DED protein family. (a) Homology model of the tandem DEDs of human FLIP (orange ribbon) depicting the main conserved residues part of the hydrophobic patch (highlighted in grey), which are essential for inter- and intra-DED–DED interactions. (b) Sequence alignment of residues that form part of the hydrophobic patches, which are highly conserved amongst DED proteins. (c) Homology model of human FLIP DEDs (orange ribbon) with a close-up view of the two E/D-RxDL motifs. Only residues that form the charged triad motifs (highlighted in cyan) are shown. Hydrogen bonds are highlighted in black dotted lines. (d) Sequence alignment of the E/D-RxDL motifs which are conserved amongst DED proteins
Binding modes of FLIP–FADD and procaspase 8–FADD interactions. FLIP binds to the DED of FADD using its DED2, whereas procaspase-8 binds to FADD using its DED1. The main residues that are important for the interactions are shown in grey: FLIP uses its F114 residue on the α2 helix to bind into a groove between α1/α4 helices in the DED of FADD with a reciprocal interaction from FADD H9 into the α2/α5 hydrophobic patch of FLIP; procaspase-8 uses the Y8 residue on its α1 helix to bind into the hydrophobic patch between the α2/α5 helices in FADD, with a reciprocal interaction from FADD F25 into the α1/α4 groove in procaspase-8
Emerging models of DISC assembly at the level of DED interactions
Two independent studies proposed a novel model of DISC assembly after finding that FADD is sub-stoichiometric at the DISC compared with death receptors and caspase-8, with three to five receptors and as many as nine caspase-8 molecules for every FADD molecule recruited to the complex.49, 50 DED-containing proteins interact with themselves and one another in a homotypic manner through their DEDs, so both groups proposed that one FADD molecule could recruit multiple DED-only proteins (procaspase-8, procaspase-10 or FLIP) as ‘chains’. In support of this model, formation of caspase-8 chains was observed in single cell studies using fluorescently tagged caspase-8. Such DED ‘filaments’ have been described before for caspase-8 and FADD;46, 51 however, their physiological relevance is questionable as they are generated in cells expressing supra-physiological levels of each DED protein.
By creating models of FLIP and procaspase-8 DEDs based on the published structure of vFLIP MC159,8 we used the NMR structure of FADD7 to perform docking experiments between the DEDs of the three proteins. These modelling experiments suggested that each protein pair could potentially interact in two distinct orientations, which involved mainly hydrophobic interactions between the α2/α5 surface (the aforementioned hydrophobic patch) in one DED and α1/α4 surface of the adjacent DED.48 Subsequent mutagenesis studies revealed that FLIP and procaspase-8 have differential affinities for the two available interaction surfaces of the FADD DED. FLIP preferentially binds to the α1/α4 surface of FADD’s DED, whereas procaspase-8 binds to FADD’s α2/α5 surface (Figure 4). Our analysis of the stoichiometry of the TRAIL-R2 DISC was not in agreement with the caspase chain models described above: in our study, sub-apoptotic DISC stimulation resulted in an approximate 1:1:1 ratio of FADD:caspase-8:FLIP; while at higher levels of DISC stimulation, there was more caspase-8 than FADD or FLIP, although there remained approximately one FADD molecule for every two molecules of FLIP/caspase-8.
Activation of caspase-8
The current model of procaspase-8 activation is that 53/55 kDa procaspase-8 zymogens are recruited to FADD as monomers via their DEDs leading to dimerisation of the procaspases, initially via their DEDs (Figure 5).52, 53, 54, 55, 56 Dimerisation of the caspase domains then occurs and results in conformational changes that reveal the enzymatic activity necessary for intra-molecular cleavage of the C-terminal portion of the caspase, liberating a p12 subunit (subsequently processed to the small p10 catalytic subunit) and simultaneously stabilising the dimer. Next, the 41/43 kDa caspase-8 intermediates in the dimer cleave one another in a trans-catalytic manner in the region between their DEDs and the large p18 catalytic subunit. The two molecules of p18-caspase-8 that are subsequently released associate with the two p10 subunits to form the active protease.57 These two steps are critical, as cleavage in the absence of dimerisation does not result in an active protease.58
Processing of procaspase-8. When two procaspase-8 molecules are co-recruited to the DISC via their DEDs, their caspase domains undergo conformational changes that exposes the enzymatic activity necessary for cleavage of the C-terminal portion of the caspase, liberating a p12 subunit, which is subsequently processed to the small p10 catalytic subunit. This initial processing step may occur in an inter-dimer manner between adjacent procaspase-8 dimers rather than the intra-dimer manner depicted.63 The 41/43 kDa caspase-8 intermediates cleave one another in a trans-catalytic manner in the region between their DEDs and the large p18 catalytic subunit. The two molecules of pro-caspase-8 that are subsequently released associate with the two p10 subunits to form the active protease.57 At lower levels of DISC stimulation or when FLIP is highly expressed, FLIP/caspase-8 heterodimers assemble at the DISC via interactions between their DEDs and those of FADD.48 The pseudo-caspase domain of FLIPL is able to induce the conformational change in procaspase-8’s caspase domain that is necessary to create its active site.59 The FLIPL:caspase-8 heterodimer is processed between the p18 and p12 subunits of both proteins, but is unable to be further processed owing to FLIPL’s lack of enzymatic activity, and this heterodimer is unable to activate apoptosis. In the case of FLIPS, heterodimerisation fails to activate procaspase-8 as the initial conformational change cannot take place in procaspase-8’s caspase domain
Procaspase-8 can also heterodimerise with FLIP at the DISC. In the case of FLIPS/R, heterodimerisation fails to activate procaspase-8 as the initial conformational change cannot take place in procaspase-8’s caspase domain;59 thus, FLIPS/R effectively acts in a dominant negative manner (Figure 5). For FLIPL, heterodimerisation results in an active enzyme, as the pseudo-catalytic domain of FLIPL is able to induce the conformational change in procaspase-8’s caspase domain that is necessary to create the active site (Figure 5).59 Indeed, it appears that FLIPL’s pseudo-caspase domain is more efficient at inducing the conformational change in the dimer than the caspase domain of another molecule of procaspase-8.60 The FLIP:caspase-8 heterodimer remains tethered to the DISC because the second step of activation, cleavage between the DEDs and p18-subunits cannot occur because of FLIPL’s lack of enzymatic activity, and the heterodimer is unable to activate the apoptotic cascade. However, the FLIPL:caspase-8 heterodimer’s enzymatic activity can cleave local substrates, most notably RIPK1, an important regulator of necroptosis (see below).61
Procaspase-8 dimerisation is required for formation of the active site.55, 56 However, as well as intra-dimer cleavage, procaspase-8 can be cleaved in an inter-dimeric manner (i.e., dimers acting in a trans manner).54, 115 a protein known to bind to ubiquitin, that moves caspase-8 into ubiquitin-rich foci.114 However, it is unknown whether translocation into these foci is necessary for caspase-8′s activation, and the implications this has for the observation that caspase-8 and p62 are both found in intracellular DISCs and autophagosomal membranes is unclear.103, 104 In subsequent work, the same group found that cytosolic p43 and p18 fragments of caspase-8 are degraded in a proteasome-dependent manner. In this latter study, caspase-8 was reported to be conjugated by degradative K48-linked ubiquitin chains on its p18 fragment by another E3 ligase, TRAF2.116 Rather than increasing its activation as is the case for CUL3-mediated K63-ubiquitination, TRAF2 acts to degrade the pool of activated caspase-8, decreasing the propensity of the cell to commit to apoptosis. Caspase-8-processed FLIPL has also been reported to interact with TRAF2, promoting activation of the NF-κB transcriptional pathway, but it is not yet clear whether these observations are related.117
Regulation of FLIP by the UPS
An additional layer of control of death receptor-mediated apoptosis is achieved by the ubiquitination of FLIP. Similar to the anti-apoptotic BCL-2 family member MCL-1, FLIPS is an extremely short-lived protein that is rapidly turned over through the ubiquitin-proteasome system.118 FLIPS is ubiquitinated on K192 and K195 in DED2 and, between these two lysine residues at position 193 is a serine residue which, when phosphorylated, inhibits the ubiquitination of the adjacent lysines.118, 119 Notably, mutational studies showed that ubiquitin-deficient mutants of FLIPS had increased half-lives (as expected) but were still recruited to the DISC, retaining their anti-apoptotic ability. In further work, the same lab identified PKCα/β as the key mediators of FLIPS phosphorylation on S193.119 FLIPL is less labile than FLIPS, although it too is turned over relatively rapidly, with a typical half-life of 2–3 h.118 A similar interplay between phosphorylation and ubiquitination is true for FLIPL, where ROS production induces the phosphorylation and subsequent ubiquitination and degradation of FLIPL.120 Moreover, K195 is a site of ubiquitin conjugation on FLIPL in response to hyperthermia.121 Our group has identified a role for the DNA repair protein Ku70 in regulation of FLIP ubiquitination. Ku70 forms a complex with FLIP protecting it from ubiquitination and subsequent degradation. This complex is regulated by the acetylation of Ku70 and thus can be manipulated pharmacologically by histone deacetylase inhibitors leading to rapid degradation of FLIP.122 Chang et al.123 reported that following TNFα stimulation, JNK is activated which in turn activates the E3 Ubiquitin ligase Itch, resulting in FLIPL ubiquitination and subsequent proteasomal degradation. This apparent link between FLIP and Itch was partly confirmed by other studies, including Panner et al.,124, 125 who identified a PTEN-Akt-Itch pathway controlling FLIPS stability and degradation.
DED Complexes in Mammalian Host Defence
Antiviral immunity
DED-containing protein complexes are critical for innate immune reactions and can assemble to induce apoptosis in response to viral infection, generally in a mitochondrial antiviral signalling adaptor (MAVS)-dependent manner. Typically, cytosolic viral RNA is recognised by one of the three RIG-1-like receptors (RLRs) retinoic acid-inducible gene I (RIG-1), melanoma differentiation-associated gene 5 (MDA5) or laboratory of genetics and physiology 2 (LGP2). RIG-1 and MDA5 undergo conformational changes upon binding to viral RNA, exposing an N-terminal CARD that interacts with and activates MAVS,126, 127 which in turn activates a type I interferon response.126
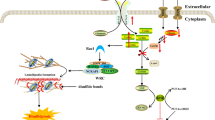
FADD and RIPK1 can interact with MAVS (Figure 7),128 and FADD- and RIPK1-deficient cells are hypersensitive to viral infection owing to an inability to induce the transcription of key antiviral genes.35 Tschopp and co-workers elucidated the mechanistic basis of these findings and found that the antiviral RIG-1 signalling pathway bears a striking resemblance to the TNFR1 pathway.64, 129 They propose that following viral infection, a complex termed the ‘TRADDosome’ (composed of TRADD, RIPK1 and FADD) forms on the mitochondrial membrane via MAVS. In this MAVS-located TRADDosome, RIPK1 can be K63 ubiquitinated by TRAF2/3 and recruit NEMO resulting in activation of IKKα and IKKβ and subsequent phosphorylation and degradation of IκB leading to NF-κB activation. Finally, via FADD, the TRADDosome can recruit procaspases-8 and -10 and FLIP, potentially inducing cell death.129
DED proteins in MAVS signaling. Following viral infection of cells, viral RNA is detected by CARD-containing RIG-1-like receptors, for example, RIG-1 and MDA5. RIG-1 binds the dsRNA, exposing its normally hidden CARD domains. K63 ubiquitin chains are conjugated to the CARDs, facilitating the assembly of a complex composed of four polyubiquitin chains and four RIG-1 molecules (not shown).200, 201 This in turn induces the formation of prion-like aggregates of MAVS, which strongly activate IRF3.202, 203 These MAVS aggregates form a platform which can recruit TRAF2, TRAF3 and TRAF6.203 TRADD also binds MAVS followed by TANK and TBK2, activating antiviral IRF3.129, 203 However, TRADD can also recruit RIPK1, FADD and caspase-8, a complex dubbed the ‘TRADDosome’. Caspase-8 cleaves RIPK1 and the resulting RIPK1 fragment can inhibit IRF3, ceasing the antiviral response.204 RIPK1 is conjugated by K63 ubiquitin chains inducing two distinct signaling pathways from the TRADDosome: firstly, NF-κB signaling through NEMO, IKKα and IKKβ, and secondly NEMO can interact with NAP1, TBK1 and IKKɛ to activate IRF3 or IRF7.205, 206, 207, 208 In addition to caspase-8, the TRADDosome can also recruit FLIP (not shown) and may under certain conditions, trigger cell death
DED proteins and the inflammasome
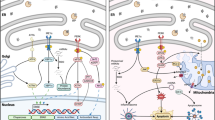
The inflammasome is a multi-protein oligomeric structure formed in macrophages and monocytes in response to inflammatory stimuli (Figure 8).130 Inflammasomes are comprised of a stimulus-specific sensor protein belonging to either the NLR, AIM2 or ALR family, the adaptor protein ASC and the inactive zymogen procaspase-1.131 Formation of the inflammasome leads to activation of caspase-1, which processes pro-IL-1β (and IL-18) to its mature form.132, 133 Different inflammasomes assemble in response to distinct stimuli, for example, the NLRP3 inflammasome forms in response to a plethora of pathogens, including influenza A,134, 135 Klebsiella pneumoniae and Staphylococcus aureus, in addition to endogenous danger signals such as ATP, uric acid crystals, nigericin and hyaluronan.131 The NLRC4 inflammasome reacts to bacterial flagellin and PrgJ, and the AIM2 inflammasome detects foreign dsDNA.136, 137 RIG-1 senses RNA viruses and forms a signaling complex with ASC, activating an inflammasome.138 Toll-like receptors (TLRs) detect pathogens by recognising pathogen membrane proteins (TLR4) or cytoplasmic nucleotides (TLR3). Full activation of inflammasomes requires two distinct stages: a ‘priming’ signal 1, for example, from either a TLR agonist or a pro-inflammatory cytokine, that activates NF-κB and upregulates pro-IL-1β expression,139, 140 followed by stimulus-specific inflammasome activation and processing of pro-IL-1β.141, 142
Involvement of DED proteins in the inflammasome. (a) Full activation of the inflammasomes requires a two-step process. Firstly, a priming signal is induced by a PAMP (such as LPS) activating a transcriptional cascade resulting in the de novo synthesis of inflammasome components such as NLRP3 and pro-IL-1β.139 Of note, subsequent work has shown that this is not always the case, as TLR-mediated priming of the NLRP3 inflammasome does not always require transcriptional upregulation of NLRP3.209, 210 (b) Assembly of the inflammasome components NLRP3, ASC and pro-caspase-1 occurs following a second signal. This can be any one of numerous different stimuli, for example, ATP activation of the P2X7 receptor, bacterial toxins, nigericin and silica.211, 212, 213 A subsequent efflux of potassium from the cell permits the components to form a functional canonical NLRP3 inflammasome where pro-caspase-1 is cleaved into its active form.214 Catalytically active caspase-1 cleaves pro-IL-1β and pro-IL-18 into their mature forms, which are then released from the cell.211 (c) Fungi and mycobacterium activate the β-glucan receptor dectin-1 resulting in the phosphorylation of cytoplasmic domain allowing the recruitment of SYK kinase.215 This elicits the formation of the non-canonical caspase-8 inflammasome, consisting of CARD9, BCL-10, MALT1, ASC and caspase-8. In this complex, active caspase-8 cleaves pro-IL-1β into its mature form which is released from the cell.216 (d) IL-1β and IL-18 can be processed directly by caspase-8 in an ASC-independent manner following ligation of members of the TNF receptor family, such as CD95, although the mechanism for this remains unclear.146 (e) The Ripoptosome forms upon loss of IAPs and can lead to activation of caspase-8, which can potentially cleave IL-1β directly or indirectly via caspase-1.150, 217
DED-containing proteins are emerging as players in inflammasome signalling. Caspase-8-dependent TLR4 signalling is critical for inflammasome assembly and IL-1β processing in glaucoma,143 and engagement of TLR3 or TLR4 can result in processing of IL-1β by caspase-8, independent of NLRP3 and caspase-1.144 Studies by Kanneganti and colleagues145 revealed that FADD and caspase-8 are obligatory for the correct priming and activation of both the canonical and non-canonical NLRP3 inflammasome, and that CD95 can induce IL-1β and IL-18 maturation in a caspase-8-dependent, but RIPK3-independent manner.146 However, work from other groups contest this: Allam et al.147 and Weng et al.148 show that caspase-8 is required only for TLR-induced inflammasome priming. A number of other studies show normal canonical NLRP3 inflammasome activation in caspase-8-deficient cells, refuting an essential role for caspase-8 in NLRP3 inflammasome activation.149, 150, 151 Together, these data suggest that caspase-8 may promote caspase-1 activity under certain conditions, but is not absolutely required for NLRP3 inflammasome activation.
FLIPL has been shown to be involved in activation of the NLRP3 and AIM2 inflammasome and directly interacts with NLRP3, AIM2 and procaspase-1.152 In contrast, FLIP decreased IL-1β generation in response to SMAC mimetics and CD95 receptor activation, indicating that its role in regulating the inflammasome is context-dependent.152
DED proteins and the adaptive immune system
It has long been known that caspase-8- or FADD-deficient T cells do not proliferate in response to T-cell receptor activation;37, 153 it had previously been thought that this was due to a defective ability to activate NF-κB, but recent work has shown that it is due to induction of necroptosis.154, 155 Rescue of T cells deficient in either caspase-8 or FADD can be achieved by simultaneously deleting RIPK1156 or by inhibiting RIPK1 pharmacologically with necrostatin-1108; furthermore, FADD−/−RIPK1−/− and Casp8−/−RIPK3−/− T cells undergo normal rates of clonal expansion following viral stimulation.97, 156, 157 Stimulation of the T-cell receptor by antigens induces the formation of the CARMA1-BCL-10-MALT1 complex which activates NF-κB.158, 159, 160, 161 Downstream of this, a complex composed of FADD, caspase-8 and FLIPL forms, which presumably prevents aberrant activation of RIPK1, blocking necroptosis and promoting cell survival and proliferation.162 TIPE2 (tumor necrosis factor-α-induced protein-8, TNFAIP8) is the newest member of the DED-containing protein family, identified as highly expressed in a murine model of spine inflammation.163, 164, 165 TIPE2−/− mice develop spontaneous fatal inflammatory disease with concomitant elevated production of pro-inflammatory cytokines, suggesting a role for TIPE2 in the immune system and, more specifically, T cells.164 Upon infection or immunisation, TIPE2−/− mice exhibit increased levels of CD8+ T cells and inflammatory cytokine production, implying that TIPE2 is a negative regulator of T-cell-mediated immunity by impeding the NF-κB and AP1 transcriptional pathways and TLR signalling in macrophages.164 Interestingly, Sun et al.165 also found that TIPE2 interacts with caspase-8, but not FLIP, in macrophages, and blockade of caspase-8 function in TIPE2−/− cells rescues the hypersensitive phenotype; however, subsequent papers have disputed this.
Therapeutically Exploiting DED Complexes
Evasion of apoptosis is a hallmark of cancer,166 but aberrant cell death is also a feature of other human pathologies such as inflammation and neurodegenerative diseases. The DED family of proteins constitute key decision makers in these processes, with the ability to switch outcomes from life to death, or to different modes of death. As such, they represent an attractive set of proteins to target therapeutically.
Death receptors such as TRAIL-R1 and TRAIL-R2 are overexpressed in many types of cancer,167 and there has been much effort to develop agents (recombinant forms of TRAIL and antibodies) that activate these receptors, particularly as they appear to selectively target malignant tissue while sparing normal cells.168 Although pre-clinical data for TRAIL receptor-targeted therapeutics were promising and these agents were well tolerated in phase I trials, they showed limited anti-cancer effects in patients when used alone or in combination with chemotherapy or proteasome inhibition (reviewed in Lemke et al.169). However, a major shortcoming of these clinical studies was that they failed to learn from the experiences with other molecularly targeted agents and were conducted in unselected patient populations.170, 171 Another limitation of first generation TRAIL receptor agonists may have been insufficient levels of receptor super-clustering; a number of second generation TRAIL agonists are now in development with novel mechanisms of action that overcome this limitation, for example, MedImmune’s multivalent ‘superagonist’, which efficiently engages and clusters TRAIL-R2.172 However, increased valency may increase the toxicity of second generation TRAIL-R agonists, and a recent phase I clinical trial with a tetravalent agonistic Nanobody targeting TRAIL-R2, TAS266, had to be halted at the lowest dose owing to hepatotoxicity. 173 The opposite approach is required for the TNFα pathway, where biologics have been developed to block TNFα itself or TNFR1, preventing downstream activation of the NF-κB pathway and/or apoptosis induction and providing effective treatment for a number of inflammatory diseases, including SLE, rheumatoid arthritis and septic shock.174, 175, 176 TNFα-induced necroptosis has been implicated in a number of pathophysiological conditions such as Crohn’s disease99 (reviewed by Linkermann and Green177); thus, TNFα blockade may prove to be therapeutically beneficial in these situations.
A number of IAP antagonists (SMAC mimetics) are currently in clinical development and have shown potential as anti-cancer agents (reviewed by Fulda178). They promote caspase activation and elicit an apoptotic response by binding to and inhibiting IAPs, which are overexpressed in many types of cancer.70, 71, 179, 180 Additionally, they activate the non-canonical NF-κB pathway through the accumulation of NIK, which is normally degraded by cIAP1.181, 182 This results in an upregulation of NF-κB target genes, including TNFα. TNFα can signal in an autocrine or paracrine manner, stimulating the assembly of TNFR1 Complex II (Figure 6) and activation of cell death via apoptosis and/or necroptosis.86, 92
As it is a potent anti-apoptotic molecule, lowering FLIP expression in malignancies could lower the threshold for cell death. We and others have reported that histone deacetylase inhibitors such as vorinostat trigger the rapid ubiquitination and degradation of FLIP, thus sensitising cells to TRAIL or chemotherapy.122, 170 Additionally, a number of chemotherapeutics and other anti-cancer agents downregulate FLIP expression via multiple mechanisms (reviewed by Safa183). As previously mentioned, the gene encoding procaspase-8 (Casp8) is silenced by methylation in several cancers, such as small cell lung cancer and neuroblastomas;184 treatment with another class of epigenetic drugs, the demethylating agents such as 5-azacytidine can reverse this effect, thereby enhancing the potential for caspase-8-mediated apoptosis.27
Conclusion and Perspective
As a result of their key roles in determining life and death outcomes, much work has focussed on the complexes formed by DED proteins. From this work, FLIP in particular has emerged as a master regulator of the signalling outputs from DED-containing complexes. It is probably for this reason that FLIP expression is regulated at multiple levels: by numerous transcription factors (such as NF-κB,185 NFAT,186 AP-1187 and c-Myc188); alternative splicing;189 mRNA translation190 and by posttranslational modifications, including its rapid turnover via the ubiquitin-proteasome system.118, 119 This exquisite level of regulation may have evolved to allow swift responses to various cellular stresses, for example, to safeguard against inappropriate activation of cell death or enhance cell death, depending on the cellular context. Biochemical and structural studies have demonstrated that DED-containing complexes are highly intricate with ubiqutination playing a key role. These complexes are also more numerous than previously appreciated, with the discovery of complexes such as the intracellular DISC and Ripoptosome and involvement of DED proteins in complexes such as the inflammasome. It is anticipated that future studies will reveal novel ways of therapeutically targeting DED protein complexes that could find clinical applications in cancers, inflammatory diseases and neurodegenerative diseases.
Abbreviations
- AIM2:
-
absent in melanoma 2
- AP-1:
-
activator protein 1
- ASC:
-
apoptosis-associated speck-like protein containing a CARD
- BCL-10:
-
B-cell lymphoma/leukemia 10
- BCL-2:
-
B-cell lymphoma 2
- BCLAF1:
-
BCL-2-associated transcription factor 1
- CARD:
-
caspase activation and recruitment domain
- CARMA1:
-
CARD-containing MAGUK protein 1
- CD95:
-
cluster of differentiation 95
- cIAP:
-
cellular inhibitor of apoptosis
- DD:
-
death domain
- DED:
-
death effector domain
- DEDD:
-
death effector domain-containing protein
- DISC:
-
death-inducing signaling complex
- FADD:
-
Fas-associated protein with death domain
- FLIP:
-
FLICE-like inhibitory protein
- HOIL-1:
-
haem-oxidized IRP2 ubiquitin ligase 1
- JNK:
-
c-Jun N-terminal kinase
- LGP2:
-
Laboratory of Genetics and Physiology 2
- MALT1:
-
mucosa-associated lymphoid tissue lymphoma translocation protein 1
- MAVS:
-
mitochondrial antiviral-signaling protein
- MCL-1:
-
myeloid cell leukemia 1
- MDA5:
-
melanoma differentiation-associated protein 5
- MLKL:
-
mixed lineage kinase-like domain
- NEMO:
-
NF-κB essential modulator
- NFAT:
-
nuclear factor of activated T cells
- NLR:
-
NOD-like receptor
- NLRP3:
-
NOD-like receptor family, pyrin domain containing 3
- PEA-15:
-
phosphoprotein enriched in astrocytes 15
- PIDD:
-
p53-induced death domain
- PTEN:
-
phosphatase and tensin homolog
- RIG-1:
-
retinoic acid-inducible gene 1
- RIPK:
-
receptor-interacting serine/threonine protein kinase
- SLE:
-
systemic lupus erythematosus
- SMAC:
-
second mitochondria-derived activator of caspases
- TAK1:
-
transforming growth factor beta-activated kinase 1
- TIPE2:
-
tumor necrosis factor-α-induced protein 8 (TNFAIP8)-like 2
- TRADD:
-
TNF receptor type 1-associated death domain
- TRAF:
-
TNF-receptor associated factor
References
Green DR, Victor B . The pantheon of the fallen: why are there so many forms of cell death? Trends Cell Biol 2012; 22: 555–556.
Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P . Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol 2014; 15: 135–147.
Tait SW, Ichim G, Green DR . Die another way - non-apoptotic mechanisms of cell death. J Cell Sci 2014; 127: 2135–2144.
Tait SW, Green DR . Mitochondria and cell signalling. J Cell Sci 2012; 125: 807–815.
Kilbride SM, Prehn JH . Central roles of apoptotic proteins in mitochondrial function. Oncogene 2013; 32: 2703–2711.
Eberstadt M, Huang B, Chen Z, Meadows RP, Ng SC, Zheng L et al. NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature 1998; 392: 941–945.
Carrington PE, Sandu C, Wei Y, Hill JM, Morisawa G, Huang T et al. The structure of FADD and its mode of interaction with procaspase-8. Mol Cell 2006; 22: 599–610.
Yang JK, Wang L, Zheng L, Wan F, Ahmed M, Lenardo MJ et al. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol Cell 2005; 20: 939–949.
Li F-Y, Jeffrey PD, Yu JW, Shi Y . Crystal structure of a viral FLIP: insights into FLIP- mediated inhibition of death receptor signaling. J Biol Chem 2006; 281: 2960–2968.
Golks A, Brenner D, Fritsch C, Krammer PH, Lavrik IN . c-FLIPR, a new regulator of death receptor-induced apoptosis. J Biol Chem 2005; 280: 14507–14513.
Degterev A, Boyce M, Yuan J . A decade of caspases. Oncogene 2003; 22: 8543–8567.
Scaffidi C, Medema JP, Krammer PH, Peter ME . FLICE is predominantly expressed as two functionally active isoforms, caspase-8/a and caspase-8/b. J Biol Chem 1997; 272: 26953–26958.
Horiuchi T, Himeji D, Tsukamoto H, Harashima S, Hashimura C, Hayashi K . Dominant expression of a novel splice variant of caspase-8 in human peripheral blood lymphocytes. Biochem Biophys Res Commun 2000; 272: 877–881.
Himeji D, Horiuchi T, Tsukamoto H, Hayashi K, Watanabe T, Harada M . Characterization of caspase-8 L: a novel isoform of caspase-8 that behaves as an inhibitor of the caspase cascade. Blood 2002; 99: 4070–4078.
Miller MA, Karacay B, Zhu X, O'Dorisio MS, Sandler AD . Caspase 8 L, a novel inhibitory isoform of caspase 8, is associated with undifferentiated neuroblastoma. Apoptosis 2006; 11: 15–24.
Vincenz C, Dixit VM . Fas-associated death domain protein interleukin-1beta-converting enzyme 2 (FLICE2), an ICE/Ced-3 homologue, is proximally involved in CD95- and p55- mediated death signaling. J Biol Chem 1997; 272: 6578–6583.
Ng PW, Porter AG, Jänicke RU . Molecular cloning and characterization of two novel pro-apoptotic isoforms of caspase-10. J Biol Chem 1999; 274: 10301–10308.
Wang H, Wang P, Sun X, Luo Y, Wang X, Ma D et al. Cloning and characterization of a novel caspase-10 isoform that activates NF-kappa B activity. Biochim Biophys Acta 2007; 1770: 1528–1537.
Wachmann K, Pop C, van Raam BJ, Drag M, Mace PD, Snipas SJ et al. Activation and specificity of human caspase-10. Biochemistry 2010; 49: 8307–8315.
Wang J, Chun HJ, Wong W, Spencer DM, Lenardo MJ . Caspase-10 is an initiator caspase in death receptor signaling. Proc Natl Acad Sci USA 2001; 98: 13884–13888.
Kischkel FC, Lawrence DA, Tinel A, LeBlanc H, Virmani A, Schow P et al. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem 2001; 276: 46639–46646.
Sprick MR, Rieser E, Stahl H, Grosse-Wilde A, Weigand MA, Walczak H . Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J 2002; 21: 4520–4530.
Fischer U, Stroh C, Schulze-Osthoff K . Unique and overlap** substrate specificities of caspase-8 and caspase-10. Oncogene 2006; 25: 152–159.
Bae S, Ha TS, Yoon Y, Lee J, Cha HJ, Yoo H et al. Genome-wide screening and identification of novel proteolytic cleavage targets of caspase-8 and -10 in vitro. Int J Mol Med 2008; 21: 381–386.
Reed JC, Doctor K, Rojas A, Zapata JM, Stehlik C, Fiorentino L et al. Comparative analysis of apoptosis and inflammation genes of mice and humans. Genome Res 2003; 13: 1376–1388.
Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA et al. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med 2000; 6: 529–535.
Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N . Loss of caspase-8 expression in highly malignant human neuroblastoma cells correlates with resistance to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res 2000; 60: 4315–4319.
Mühlethaler-Mottet A, Flahaut M, Bourloud KB, Nardou K, Coulon A, Liberman J et al. Individual caspase-10 isoforms play distinct and opposing roles in the initiation of death receptor-mediated tumour cell apoptosis. Cell Death Dis 2011; 2: e125.
Sakamaki K, Shimizu K, Iwata H, Imai K, Satou Y, Funayama N et al. The apoptotic initiator caspase-8: its functional ubiquity and genetic diversity during animal evolution. Mol Biol Evol 2014; 31: 3282–3301.
Quistad SD, Stotland A, Barott KL, Smurthwaite CA, Hilton BJ, Grasis JA et al. Evolution of TNF-induced apoptosis reveals 550 My of functional conservation. Proc Natl Acad Sci USA 2014; 111: 9567–9572.
Scaffidi C, Volkland J, Blomberg I, Hoffmann I, Krammer PH, Peter ME . Phosphorylation of FADD/ MORT1 at serine 194 and association with a 70-kDa cell cycle-regulated protein kinase. J Immunol 2000; 164: 1236–1242.
Zhang J, Kabra NH, Cado D, Kang C, Winoto A . FADD-deficient T cells exhibit a disaccord in regulation of the cell cycle machinery. J Biol Chem 2001; 276: 29815–29818.
Chen G, Bhojani MS, Heaford AC, Chang DC, Laxman B, Thomas DG et al. Phosphorylated FADD induces NF-kappaB, perturbs cell cycle, and is associated with poor outcome in lung adenocarcinomas. Proc Natl Acad Sci USA 2005; 102: 12507–12512.
Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem 2005; 280: 20722–20729.
Balachandran S, Thomas E, Barber GN . A FADD-dependent innate immune mechanism in mammalian cells. Nature 2004; 432: 401–405.
Ma Y, Liu H, Tu-Rapp H, Thiesen HJ, Ibrahim SM, Cole SM et al. Fas ligation on macrophages enhances IL-1R1-Toll-like receptor 4 signaling and promotes chronic inflammation. Nat Immunol 2004; 5: 380–387.
Zhang J, Cado D, Chen A, Kabra NH, Winoto A . Fas-mediated apoptosis and activation- induced T-cell proliferation are defective in mice lacking FADD/Mort1. Nature 1998; 392: 296–300.
Chan FK, Chun HJ, Zheng L, Siegel RM, Bui KL, Lenardo MJ . A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 2000; 288: 2351–2354.
Siegel RM, Frederiksen JK, Zacharias DA, Chan FK, Johnson M, Lynch D et al. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science 2000; 288: 2354–2357.
Scott FL, Stec B, Pop C, Dobaczewska MK, Lee JJ, Monosov E et al. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature 2009; 457: 1019–1022.
Martin DA, Zheng L, Siegel RM, Huang B, Fisher GH, Wang J et al. Defective CD95/APO-1/Fas signal complex formation in the human autoimmune lymphoproliferative syndrome, type Ia. Proc Natl Acad Sci USA 1999; 96: 4552–4557.
Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, Park AY et al. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat Struct Mol Biol 2010; 17: 1324–1329.
Esposito D, Sankar A, Morgner N, Robinson CV, Rittinger K, Driscoll PC . Solution NMR investigation of the CD95/FADD homotypic death domain complex suggests lack of engagement of the CD95 C terminus. Structure 2010; 18: 1378–1390.
Sandu C, Morisawa G, Wegorzewska I, Huang T, Arechiga AF, Hill JM et al. FADD self- association is required for stable interaction with an activated death receptor. Cell Death Differ 2006; 13: 2052–2061.
Siegel RM, Muppidi JR, Sarker M, Lobito A, Jen M, Martin D et al. SPOTS: signaling protein oligomeric transduction structures are early mediators of death receptor-induced apoptosis at the plasma membrane. J Cell Biol 2004; 167: 735–744.
Garvey TL, Bertin J, Siegel RM, Wang GH, Lenardo MJ, Cohen JI . Binding of FADD and caspase-8 to molluscum contagiosum virus MC159 v-FLIP is not sufficient for its antiapoptotic function. J Virol 2002; 76: 697–706.
Ueffing N, Keil E, Freund C, Kühne R, Schulze-Osthoff K, Schmitz I . Mutational analyses of c-FLIPR, the only murine short FLIP isoform, reveal requirements for DISC recruitment. Cell Death Differ 2008; 15: 773–782.
Majkut J, Sgobba M, Holohan C, Crawford N, Logan AE, Kerr E et al. Differential affinity of FLIP and procaspase 8 for FADD's DED binding surfaces regulates DISC assembly. Nat Commun 2014; 5: 3350.
Schleich K, Warnken U, Fricker N, Öztürk S, Richter P, Kammerer K et al. Stoichiometry of the CD95 death-inducing signaling complex: experimental and modeling evidence for a death effector domain chain model. Mol Cell 2012; 47: 306–319.
Dickens LS, Boyd RS, Jukes-Jones R, Hughes MA, Robinson GL, Fairall L et al. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Mol Cell 2012; 47: 291–305.
Siegel RM, Martin DA, Zheng L, Ng SY, Bertin J, Cohen J et al. Death-effector filaments: novel cytoplasmic structures that recruit caspases and trigger apoptosis. J Cell Biol 1998; 141: 1243–1253.
Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM . An induced proximity model for caspase-8 activation. J Biol Chem 1998; 273: 2926–2930.
Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM et al. A unified model for apical caspase activation. Mol Cell 2003; 11: 529–541.
Pop C, Fitzgerald P, Green DR, Salvesen GS . Role of proteolysis in caspase-8 activation and stabilization. Biochemistry 2007; 46: 4398–4407.
Keller N, Mares J, Zerbe O, Grütter MG . Structural and biochemical studies on procaspase-8: new insights on initiator caspase activation. Structure 2009; 17: 438–448.
Keller N, Grütter MG, Zerbe O . Studies of the molecular mechanism of caspase-8 activation by solution NMR. Cell Death Differ 2010; 17: 710–718.
Medema JP, Scaffidi C, Kischkel FC, Shevchenko A, Mann M, Krammer PH et al. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC). EMBO J 1997; 16: 2794–2804.
Oberst A, Pop C, Tremblay AG, Blais V, Denault JB, Salvesen GS et al. Inducible dimerization and inducible cleavage reveal a requirement for both processes in caspase-8 activation. J Biol Chem 2010; 285: 16632–16642.
Yu JW, Jeffrey PD, Shi Y . Mechanism of procaspase-8 activation by c-FLIPL. Proc Natl Acad Sci USA 2009; 106: 8169–8174.
Boatright KM, Deis C, Denault J-B, Sutherlin DP, Salvesen GS . Activation of caspases-8 and -10 by FLIP(L). Biochem J 2004; 382: 651–657.
Pop C, Oberst A, Drag M, van Raam BJ, Riedl SJ, Green DR et al. FLIP(L) induces caspase 8 activity in the absence of interdomain caspase 8 cleavage and alters substrate specificity. Biochem J 2011; 433: 447–457.
Chang DW, **ng Z, Capacio VL, Peter ME, Yang X . Interdimer processing mechanism of procaspase-8 activation. EMBO J 2003; 22: 4132–4142.
Kallenberger SM, Beaudouin J, Claus J, Fischer C, Sorger PK, Legewie S et al. Intra- and interdimeric caspase-8 self-cleavage controls strength and timing of CD95-induced apoptosis. Sci Signal 2014; 7: ra23.
Micheau O, Tschopp J . Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 2003; 114: 181–190.
Tada K, Okazaki T, Sakon S, Kobarai T, Kurosawa K, Yamaoka S et al. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-kappa B activation and protection from cell death. J Biol Chem 2001; 276: 36530–36534.
Hsu H, Shu HB, Pan MG, Goeddel DV . TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 1996; 84: 299–308.
Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV . The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 1995; 83: 1243–1252.
Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L et al. c- IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J 2010; 29: 4198–4209.
Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci USA 2008; 105: 11778–11783.
Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell 2008; 30: 689–700.
Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 2007; 131: 682–693.
Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K et al. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF- kappaB activation. J Biol Chem 2008; 283: 24295–24299.
Ea CK, Deng L, **a ZP, Pineda G, Chen ZJ . Activation of IKK by TNFalpha requires site- specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell 2006; 22: 245–257.
Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol 2009; 11: 123–132.
Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 2009; 36: 831–844.
Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 2011; 471: 591–596.
Moulin M, Anderton H, Voss AK, Thomas T, Wong WW, Bankovacki A et al. IAPs limit activation of RIP kinases by TNF receptor 1 during development. EMBO J 2012; 31: 1679–1691.
Wong WW, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J . RIPK1 is not essential for TNFR1-induced activation of NF-kappaB. Cell Death Differ 2010; 17: 482–487.
Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S-I et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 2011; 471: 633–636.
Ikeda F, Deribe YL, Skånland SS, Stieglitz B, Grabbe C, Franz-Wachtel M et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 2011; 471: 637–641.
Wright A, Reiley WW, Chang M, ** W, Lee AJ, Zhang M et al. Regulation of early wave of germ cell apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev Cell 2007; 13: 705–716.
Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–1123.
Sun L, Wang H, Wang Z, He S, Chen S, Liao D et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012; 148: 213–227.
Cai Z, Jitkaew S, Zhao J, Chiang HC, Choksi S, Liu J et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol 2014; 16: 55–65.
Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 2011; 471: 363–367.
Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 2011; 43: 449–463.
Robinson N, McComb S, Mulligan R, Dudani R, Krishnan L, Sad S . Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat Immunol 2012; 13: 954–962.
Kim JH, Kim SJ, Lee IS, Lee MS, Uematsu S, Akira S et al. Bacterial endotoxin induces the release of high mobility group box 1 via the IFN-beta signaling pathway. J Immunol 2009; 182: 2458–2466.
Kaczmarek A, Vandenabeele P, Krysko DV . Necroptosis: the release of damage- associated molecular patterns and its physiological relevance. Immunity 2013; 38: 209–223.
Cullen SP, Henry CM, Kearney CJ, Logue SE, Feoktistova M, Tynan GA et al. Fas/CD95- induced chemokines can serve as ‘Find-Me’ signals for apoptotic cells. Mol Cell 2013; 49: 1034–1048.
Kearney CJ, Cullen SP, Tynan GA, Henry CM, Clancy D, Lavelle EC et al. Necroptosis suppresses inflammation via termination of TNF- or LPS-induced cytokine and chemokine production. Cell Death Differ 2015; 22: 1313–1327.
Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 2011; 43: 432–448.
Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008; 4: 313–321.
Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL et al. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 1998; 9: 267–276.
Yeh WC, Pompa JL, McCurrach ME, Shu HB, Elia AJ, Shahinian A et al. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science 1998; 279: 1954–1958.
Yeh WC, Itie A, Elia AJ, Ng M, Shu HB, Wakeham A et al. Requirement for Casper (c- FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity 2000; 12: 633–642.
Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 2011; 471: 368–372.
Kovalenko A, Kim JC, Kang TB, Rajput A, Bogdanov K, Dittrich-Breiholz O et al. Caspase-8 deficiency in epidermal keratinocytes triggers an inflammatory skin disease. J Exp Med 2009; 206: 2161–2177.
Günther C, Martini E, Wittkopf N, Amann K, Weigmann B, Neumann H et al. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature 2011; 477: 335–339.
Weinlich R, Oberst A, Dillon CP, Janke LJ, Milasta S, Lukens JR et al. Protective roles for caspase-8 and cFLIP in adult homeostasis. Cell Rep 2013; 5: 340–348.
Kang TB, Ben-Moshe T, Varfolomeev EE, Pewzner-Jung Y, Yogev N, Jurewicz A et al. Caspase-8 serves both apoptotic and nonapoptotic roles. J Immunol 2004; 173: 2976–2984.
Dillon CP, Oberst A, Weinlich R, Janke LJ, Kang TB, Ben-Moshe T et al. Survival function of the FADD-CASPASE-8-cFLIP(L) complex. Cell Rep 2012; 1: 401–407.
Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)- mediated caspase-8 activation and apoptosis. J Biol Chem 2012; 287: 12455–12468.
Huang S, Okamoto K, Yu C, Sinicrope FA . p62/sequestosome-1 up-regulation promotes ABT-263-induced caspase-8 aggregation/activation on the autophagosome. J Biol Chem 2013; 288: 33654–33666.
White E . Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 2012; 12: 401–410.
Maiuri MC, Zalckvar E, Kimchi A, Kroemer G . Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8: 741–752.
Laussmann MA, Passante E, Düssmann H, Rauen JA, Würstle ML, Delgado ME et al. Proteasome inhibition can induce an autophagy-dependent apical activation of caspase-8. Cell Death Differ 2011; 18: 1584–1597.
Bell BD, Leverrier S, Weist BM, Newton RH, Arechiga AF, Luhrs KA et al. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc Natl Acad Sci USA 2008; 105: 16677–16682.
Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 2004; 304: 1500–1502.
Lin Y, Devin A, Rodriguez Y, Liu ZG . Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev 1999; 13: 2514–2526.
Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol 2009; 11: 1355–1362.
Lamy L, Ngo VN, Emre NC, Shaffer AL, Yang Y, Tian E et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell 2013; 23: 435–449.
Kasof GM, Goyal L, White E . Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol Cell Biol 1999; 19: 4390–4404.
** Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 2009; 137: 721–735.
Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW . Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol 2004; 24: 8055–8068.
Gonzalvez F, Lawrence D, Yang B, Yee S, Pitti R, Marsters S et al. TRAF2 Sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Mol Cell 2012; 48: 888–899.
Kataoka T, Tschopp J . N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol Cell Biol 2004; 24: 2627–2636.
Poukkula M, Kaunisto A, Hietakangas V, Denessiouk K, Katajamäki T, Johnson MS et al. Rapid turnover of c-FLIPshort is determined by its unique C-terminal tail. J Biol Chem 2005; 280: 27345–27355.
Kaunisto A, Kochin V, Asaoka T, Mikhailov A, Poukkula M, Meinander A et al. PKC-mediated phosphorylation regulates c-FLIP ubiquitylation and stability. Cell Death Differ 2009; 16: 1215–1226.
Wilkie-Grantham RP, Matsuzawa S, Reed JC . Novel phosphorylation and ubiquitination sites regulate reactive oxygen species-dependent degradation of anti-apoptotic c-FLIP protein. J Biol Chem 2013; 288: 12777–12790.
Song X, Kim SY, Zhou Z, Lagasse E, Kwon YT, Lee YJ . Hyperthermia enhances mapatumumab-induced apoptotic death through ubiquitin-mediated degradation of cellular FLIP(long) in human colon cancer cells. Cell Death Dis 2013; 4: e577.
Kerr E, Holohan C, Mclaughlin KM, Majkut J, Dolan S, Redmond K et al. Identification of an acetylation-dependant Ku70/FLIP complex that regulates FLIP expression and HDAC inhibitor-induced apoptosis. Cell Death Differ 2012; 19: 1317–1327.
Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell 2006; 124: 601–613.
Panner A, Crane CA, Weng C, Feletti A, Parsa AT, Pieper RO . A novel PTEN-dependent link to ubiquitination controls FLIPS stability and TRAIL sensitivity in glioblastoma multiforme. Cancer Res 2009; 69: 7911–7916.
Panner A, Crane CA, Weng C, Feletti A, Fang S, Parsa AT et al. Ubiquitin-specific protease 8 links the PTEN-Akt-AIP4 pathway to the control of FLIPS stability and TRAIL sensitivity in glioblastoma multiforme. Cancer Res 2010; 70: 5046–5053.
Seth RB, Sun L, Ea CK, Chen ZJ . Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 2005; 122: 669–682.
Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA 2007; 104: 582–587.
Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 2005; 6: 981–988.
Michallet MC, Meylan E, Ermolaeva MA, Vazquez J, Rebsamen M, Curran J et al. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity 2008; 28: 651–661.
Martinon F, Burns K, Tschopp J . The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 2002; 10: 417–426.
Rathinam VA, Vanaja SK, Fitzgerald KA . Regulation of inflammasome signaling. Nat Immunol 2012; 13: 333–342.
Lamkanfi M, Dixit VM . Inflammasomes: guardians of cytosolic sanctity. Immunol Rev 2009; 227: 95–105.
Strowig T, Henao-Mejia J, Elinav E, Flavell R . Inflammasomes in health and disease. Nature 2012; 481: 278–286.
Thomas PG, Dash P, Aldridge JR, Ellebedy AH, Reynolds C, Funk AJ et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 2009; 30: 566–575.
Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 2009; 30: 556–565.
Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES . AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 2009; 458: 509–513.
Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 2009; 323: 1057–1060.
Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol 2010; 11: 63–69.
Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol 2009; 183: 787–791.
Franchi L, Eigenbrod T, Nuñez G . Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J Immunol 2009; 183: 792–796.
Latz E, **ao TS, Stutz A . Activation and regulation of the inflammasomes. Nat Rev Immunol 2013; 13: 397–411.
Lamkanfi M, Dixit VM . Mechanisms and functions of inflammasomes. Cell 2014; 157: 1013–1022.
Chi W, Li F, Chen H, Wang Y, Zhu Y, Yang X et al. Caspase-8 promotes NLRP1/NLRP3 inflammasome activation and IL-1β production in acute glaucoma. Proc Natl Acad Sci USA 2014; 111: 11181–11186.
Maelfait J, Vercammen E, Janssens S, Schotte P, Haegman M, Magez S et al. Stimulation of Toll-like receptor 3 and 4 induces interleukin-1beta maturation by caspase-8. J Exp Med 2008; 205: 1967–1973.
Gurung P, Anand PK, Malireddi RK, Vande Walle L, Van Opdenbosch N, Dillon CP et al. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J Immunol 2014; 192: 1835–1846.
Bossaller L, Chiang PI, Schmidt-Lauber C, Ganesan S, Kaiser WJ, Rathinam VA et al. Cutting edge: FAS (CD95) mediates noncanonical IL-1β and IL-18 maturation via caspase-8 in an RIP3-independent manner. J Immunol 2012; 189: 5508–5512.
Allam R, Lawlor KE, Yu EC, Mildenhall AL, Moujalled DM, Lewis RS et al. Mitochondrial apoptosis is dispensable for NLRP3 inflammasome activation but non- apoptotic caspase-8 is required for inflammasome priming. EMBO Rep 2014; 15: 982–990.
Weng D, Marty-Roix R, Ganesan S, Proulx MK, Vladimer GI, Kaiser WJ et al. Caspase-8 and RIP kinases regulate bacteria-induced innate immune responses and cell death. Proc Natl Acad Sci USA 2014; 111: 7391–7396.
Ganesan S, Rathinam VA, Bossaller L, Army K, Kaiser WJ, Mocarski ES et al. Caspase-8 modulates dectin-1 and complement receptor 3-driven IL-1β production in response to β-glucans and the fungal pathogen, Candida albicans. J Immunol 2014; 193: 2519–2530.
Lawlor KE, Khan N, Mildenhall A, Gerlic M, Croker BA, D'Cruz AA et al. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat Commun 2015; 6: 6282.
Philip NH, Dillon CP, Snyder AG, Fitzgerald P, Wynosky-Dolfi MA, Zwack EE et al. Caspase-8 mediates caspase-1 processing and innate immune defense in response to bacterial blockade of NF-κB and MAPK signaling. Proc Natl Acad Sci USA 2014; 111: 7385–7390.
Wu YH, Kuo WC, Wu YJ, Yang KT, Chen ST, Jiang ST et al. Participation of c-FLIP in NLRP3 and AIM2 inflammasome activation. Cell Death Differ 2014; 21: 451–461.
Salmena L, Hakem R . Caspase-8 deficiency in T cells leads to a lethal lymphoinfiltrative immune disorder. J Exp Med 2005; 202: 727–732.
Lu JV, Weist BM, van Raam BJ, Marro BS, Nguyen LV, Srinivas P et al. Complementary roles of Fas-associated death domain (FADD) and receptor interacting protein kinase-3 (RIPK3) in T-cell homeostasis and antiviral immunity. Proc Natl Acad Sci USA 2011; 108: 15312–15317.
Ch'en IL, Tsau JS, Molkentin JD, Komatsu M, Hedrick SM . Mechanisms of necroptosis in T cells. J Exp Med 2011; 208: 633–641.
Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J . Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature 2011; 471: 373–376.
Osborn SL, Diehl G, Han SJ, Xue L, Kurd N, Hsieh K et al. Fas-associated death domain (FADD) is a negative regulator of T-cell receptor-mediated necroptosis. Proc Natl Acad Sci USA 2010; 107: 13034–13039.
Ruland J, Duncan GS, Elia A, del Barco Barrantes I, Nguyen L, Plyte S et al. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell 2001; 104: 33–42.
Gaide O, Favier B, Legler DF, Bonnet D, Brissoni B, Valitutti S et al. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat Immunol 2002; 3: 836–843.
Ruefli-Brasse AA, French DM, Dixit VM . Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science 2003; 302: 1581–1584.
Kawadler H, Gantz MA, Riley JL, Yang X . The paracaspase MALT1 controls caspase-8 activation during lymphocyte proliferation. Mol Cell 2008; 31: 415–421.
Misra RS, Russell JQ, Koenig A, Hinshaw-Makepeace JA, Wen R, Wang D et al. Caspase-8 and c-FLIPL associate in lipid rafts with NF-kappaB adaptors during T cell activation. J Biol Chem 2007; 282: 19365–19374.
Carmody RJ, Hilliard B, Maguschak K, Chodosh LA, Chen YH . Genomic scale profiling of autoimmune inflammation in the central nervous system: the nervous response to inflammation. J Neuroimmunol 2002; 133: 95–107.
Sun H, Gong S, Carmody RJ, Hilliard A, Li L, Sun J et al. TIPE2, a negative regulator of innate and adaptive immunity that maintains immune homeostasis. Cell 2008; 133: 415–426.
Zhang X, Wang J, Fan C, Li H, Sun H, Gong S et al. Crystal structure of TIPE2 provides insights into immune homeostasis. Nat Struct Mol Biol 2009; 16: 89–90.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
McLornan DP, Barrett HL, Cummins R, McDermott U, McDowell C, Conlon SJ et al. Prognostic significance of TRAIL signaling molecules in stage II and III colorectal cancer. Clin Cancer Res 2010; 16: 3442–3451.
Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest 1999; 104: 155–162.
Lemke J, Karstedt von S, Zinngrebe J, Walczak H . Getting TRAIL back on track for cancer therapy. Cell Death Differ 2014; 21: 1350–1364.
Riley JS, Hutchinson R, McArt DG, Crawford N, Holohan C, Paul I et al. Prognostic and therapeutic relevance of FLIP and procaspase-8 overexpression in non-small cell lung cancer. Cell Death Dis 2013; 4: e951.
Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K et al. Death- receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med 2007; 13: 1070–1077.
Swers JS, Grinberg L, Wang L, Feng H, Lekstrom K, Carrasco R et al. Multivalent scaffold proteins as superagonists of TRAIL receptor 2-induced apoptosis. Mol Cancer Ther 2013; 12: 1235–1244.
Papadopoulos KP, Isaacs R, Bilic S, Kentsch K, Huet HA, Hofmann M et al. Unexpected hepatotoxicity in a phase I study of TAS266, a novel tetravalent agonistic Nanobody targeting the DR5 receptor. Cancer Chemother Pharmacol 2015; 75: 887–895.
Tracey KJ, Fong Y, Hesse DG, Manogue KR, Lee AT, Kuo GC et al. Anti-cachectin/TNF monoclonal antibodies prevent septic shock during lethal bacteraemia. Nature 1987; 330: 662–664.
Williams RO, Feldmann M, Maini RN . Anti-tumor necrosis factor ameliorates joint disease in murine collagen-induced arthritis. Proc Natl Acad Sci USA 1992; 89: 9784–9788.
Elliott MJ, Maini RN, Feldmann M, Long-Fox A, Charles P, Bijl H et al. Repeated therapy with monoclonal antibody to tumour necrosis factor alpha (cA2) in patients with rheumatoid arthritis. Lancet 1994; 344: 1125–1127.
Linkermann A, Green DR . Necroptosis. N Engl J Med 2014; 370: 455–465.
Fulda S . Smac mimetics as IAP antagonists. Semin Cell Dev Biol 2015; 39: 132–138.
Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 2007; 131: 669–681.
Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J et al. Autocrine TNFalpha signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell 2007; 12: 445–456.
Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng P-H, Keats JJ, Wang H et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol 2008; 9: 1364–1370.
Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol 2008; 9: 1371–1378.
Safa A, Pollock KE . Targeting the anti-apoptotic protein c-FLIP for cancer therapy. Cancers 2011; 3: 1639–1671.
Casicano I, Banelli B, Croce M, De Ambrosis A, di Vinci A, Gelvi I et al. Caspase-8 gene expression in neuroblastoma. Ann N Y Acad Sci 2004; 1028: 157–167.
Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J . NF-kappaB signals induce the expression of c-FLIP. Mol Cell Biol 2001; 21: 5299–5305.
Ueffing N, Schuster M, Keil E, Schulze-Osthoff K, Schmitz I . Up-regulation of c-FLIP short by NFAT contributes to apoptosis resistance of short-term activated T cells. Blood 2008; 112: 690–698.
Li W, Zhang X, Olumi AF . MG-132 sensitizes TRAIL-resistant prostate cancer cells by activating c-Fos/c-Jun heterodimers and repressing c-FLIP(L). Cancer Res 2007; 67: 2247–2255.
Ricci MS, ** Z, Dews M, Yu D, Thomas-Tikhonenko A, Dicker DT et al. Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol Cell Biol 2004; 24: 8541–8555.
Park SJ, Kim YY, Ju JW, Han BG, Park SI, Park BJ . Alternative splicing variants of c-FLIP transduce the differential signal through the Raf or TRAF2 in TNF-induced cell proliferation. Biochem Biophys Res Commun 2001; 289: 1205–1210.
Fan S, Li Y, Yue P, Khuri FR, Sun S-Y . The eIF4E/eIF4G interaction inhibitor 4EGI-1 augments TRAIL-mediated apoptosis through c-FLIP Down-regulation and DR5 induction independent of inhibition of cap-dependent protein translation. Neoplasia 2010; 12: 346–356.
Twomey EC, Wei Y . High-definition NMR structure of PED/PEA-15 death effector domain reveals details of key polar side chain interactions. Biochem Biophys Res Commun 2012; 424: 141–146.
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y . The I-TASSER Suite: protein structure and function prediction. Nat Methods 2015; 12: 7–8.
O'Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT . Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr Biol 2007; 17: 418–424.
Wang L, Du F, Wang X . TNF-alpha induces two distinct caspase-8 activation pathways. Cell 2008; 133: 693–703.
Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci USA 2012; 109: 5322–5327.
Wang H, Sun L, Su L, Rizo J, Liu L, Wang L-F et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell 2014; 54: 133–146.
Su L, Quade B, Wang H, Sun L, Wang X, Rizo J . A plug release mechanism for membrane permeation by MLKL. Structure 2014; 22: 1489–1500.
Hildebrand JM, Tanzer MC, Lucet IS, Young SN, Spall SK, Sharma P et al. Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc Natl Acad Sci USA 2014; 111: 15072–15077.
Dondelinger Y, Declercq W, Montessuit S, Roelandt R, Goncalves A, Bruggeman I et al. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep 2014; 7: 971–981.
Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 2010; 141: 315–330.
Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 2012; 36: 959–973.
Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ . MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 2011; 146: 448–461.
Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. eLife 2014; 3: e01489.
Rajput A, Kovalenko A, Bogdanov K, Yang SH, Kang TB, Kim J-C et al. RIG-I RNA helicase activation of IRF3 transcription factor is negatively regulated by caspase-8- mediated cleavage of the RIP1 protein. Immunity 2011; 34: 340–351.
Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 2003; 4: 491–496.
Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J . Triggering the interferon antiviral response through an IKK-related pathway. Science 2003; 300: 1148–1151.
Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H et al. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med 2004; 199: 1641–1650.
Perry AK, Chow EK, Goodnough JB, Yeh WC, Cheng G . Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J Exp Med 2004; 199: 1651–1658.
Juliana C, Fernandes-Alnemri T, Kang S, Farias A, Qin F, Alnemri ES . Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J Biol Chem 2012; 287: 36617–36622.
Fernandes-Alnemri T, Kang S, Anderson C, Sagara J, Fitzgerald KA, Alnemri ES . Cutting edge: TLR signaling licenses IRAK1 for rapid activation of the NLRP3 inflammasome. J Immunol 2013; 191: 3995–3999.
Perregaux D, Gabel CA . Interleukin-1 beta maturation and release in response to ATP and nigericin. Evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J Biol Chem 1994; 269: 15195–15203.
Ferrari D, Chiozzi P, Falzoni S, Dal Susino M, Melchiorri L, Baricordi OR et al. Extracellular ATP triggers IL-1 beta release by activating the purinergic P2Z receptor of human macrophages. J Immunol 1997; 159: 1451–1458.
Kahlenberg JM, Lundberg KC, Kertesy SB, Qu Y, Dubyak GR . Potentiation of caspase-1 activation by the P2X7 receptor is dependent on TLR signals and requires NF-kappaB- driven protein synthesis. J Immunol 2005; 175: 7611–7622.
Muñoz-Planillo R, Kuffa P, Martínez-Colón G, Smith BL, Rajendiran TM, Nuñez G . K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 2013; 38: 1142–1153.
Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, Schweighoffer E et al. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity 2005; 22: 507–517.
Gringhuis SI, Kaptein TM, Wevers BA, Theelen B, van der Vlist M, Boekhout T et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1β via a noncanonical caspase-8 inflammasome. Nat Immunol 2012; 13: 246–254.
Moriwaki K, Bertin J, Gough PJ, Chan FK . A RIPK3-caspase 8 complex mediates atypical pro-IL-1β processing. J Immunol 2015; 194: 1938–1944.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by P Ekert
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Riley, J., Malik, A., Holohan, C. et al. DED or alive: assembly and regulation of the death effector domain complexes. Cell Death Dis 6, e1866 (2015). https://doi.org/10.1038/cddis.2015.213
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2015.213
- Springer Nature Limited