Abstract
Gene polymorphism of acetaldehyde dehydrogenase 2 (ALDH2), a key enzyme for alcohol metabolism in humans, can affect catalytic activity. The ALDH2 Glu504Lys mutant allele has a high-frequency distribution in East Asian populations and has been demonstrated to be associated with an increased risk of cardiovascular disease, stroke, and tumors. Available evidence suggests that the evolution of the ALDH2 gene has been influenced by multiple factors. Random mutations produce Glu504Lys, and genetic drift alters the frequency of this allele; additionally, environmental factors such as hepatitis B virus infection and high-elevation hypoxia affect its frequency through selective effects, ultimately resulting in a high frequency of this allele in East Asian populations. Here, the origin, selection, and spread of the ALDH2 Glu504Lys allele are discussed, and an outlook for further research is proposed to realize a precision medical strategy based on the genetic and environmental variations in ALDH2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The acetaldehyde dehydrogenase (ALDH) superfamily consists of enzymes that catalyze the oxidation of acetaldehyde to acidic metabolites, participating in a variety of physiological and pathological processes (Jackson et al. 2011). Nineteen isoenzymes have been identified; the most active one is acetaldehyde dehydrogenase 2 (ALDH2), a 56-kDa tetramer that plays an important role in alcohol metabolism, converting acetaldehyde, an intermediate of alcohol metabolism, to acetic acid (Vasiliou et al. 1999). However, a G to A mutation at rs671 in the ALDH2 gene causes replacement of glutamate with lysine at residue 504, resulting in a reduction in ALDH2 activity, allowing acetaldehyde to accumulate in the body after alcohol consumption, with adverse effects such as flushing and tachycardia (Matsumura et al. 2019).
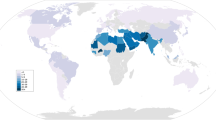
The frequency of this variant of ALDH2 (ALDH2 Glu504Lys) is low in caucasian, whereas the frequency is 30–50% in East Asian populations, especially in Chinese, Korean and Japanese (Goedde et al. 1992). ALDH2 has been shown to play an important role in a variety of diseases. According to our previous studies, mitochondrial ALDH2 acts as endogenous protection in the heart, and it is closely associated with the development of several cardiovascular diseases (Li et al. 2019). Our studies have demonstrated that patients with ALDH2 Glu504Lys variant are more inclined to suffer from coronary artery lesions (Xu et al. 2018) and displayed a higher risk of develo** poor coronary collateral circulation (Liu et al. 2015). Downregulation of ALDH2 in the mitochondria can also lead to cardiomyocyte apoptosis after myocardial infarction in mouse models (Sun et al. 2014). In addition, we have found that ALDH2 deficiency can aggravate energy metabolism disturbance and diastolic dysfunction in diabetic mice (Wang et al. 2016). Moreover, it has been shown that the ALDH2 Glu504Lys variant attenuates bioconversion of nitroglycerin, thus reducing its therapeutic effect on coronary artery disease (Li et al. It is now generally accepted that humans originated in Africa (Ingman et al. 2000). Approximately 100,000 years ago, the ancestors of modern humans began to migrate out of Africa and around the world, reaching southern East Asia approximately 50,000–60,000 years ago and then spreading from south to north throughout the region (Palanichamy et al. 2004). Studies examining genetic polymorphisms on the Y chromosome and mitochondrial DNA suggested that various populations underwent multiple divisions and fusions during this migration from Africa to East Asia (Underhill et al. 2001). Since there are rare reports of ALDH2 Glu504Lys in populations of African and European ancestry, we hypothesize that this gene arose after humans migrated from Africa to the rest of the world and originated in the part of African archaic humans that settled in East Asia. Mutations arise through a random process, not an adaptation to the environment, and even the majority of mutations found in experiments are deleterious (Nei 2005). Several point mutations in the ALDH2 gene are produced, but the mutation at the rs671 locus affects enzyme activity, and it became a deleterious mutation. Genetic drift played an important role in the small breeding populations in ancient East Asia; indeed, the populations were sparsely distributed, with considerable distances among populations (Lynch et al. 2016). When a population is small, random mating between individuals causes random fluctuations in allele or haplotype frequencies, such that a certain allele or haplotype will eventually be fixed. The evolution due to genetic drift is random. The genetic variation fixed in a population is independent of the effect of natural selection, and the effect is stronger in smaller populations (Masel 2011). It has been suggested that the ALDH2 Glu504Lys allele originated from the Pai-Yuei tribe, who was distributed along the southeastern coast of China thousands of years ago (Luo et al. 2009). It has also been suggested that rather than originating in indigenous populations in the region where it now has the highest frequencies, the ALDH2 ∗ 504Lys allele was most likely carried south by Han Chinese immigrants from central China (Li et al. 2009). Although these hypotheses are lack of strong evidence, they suggest that the mutation probably originated in a specific population, namely, an ancient tribe with a small population and the lack of genetic exchange, after which a stable frequency of the mutated gene gradually developed after several generations of genetic drift. In a more extreme case, the founder effect, in which some of the genes of a few ancestors of the population gradually reach high levels due to genetic drift, may occur if the population is completely isolated, without any gene exchange with other populations (Zlotogora 1994). Although the generation of the ALDH2 Glu504Lys allele can be explained by random mutations and genetic drift, these effects on gene polymorphism are very limited and confined to small populations. Hence, there may be some selective forces to maintain its high frequency in East Asia. Indeed, some studies on ALDH2 genetic polymorphism have indicated that the ALDH2 ∗ 504Lys allele had some compensating advantage because they tended to drink less alcohol or had some other advantages (Li et al. 2009; Goldman and Enoch 1990; Lin and Cheng 2002), as in the case of the gene that causes sickle cell anemia conferring a compensating advantage of a certain level of resistance to malaria. Although this issue is currently unresolved, the available evidence suggests that multiple factors may be involved in this process. Selection theory suggests that natural selection plays a major role in the generation and maintenance of genetic polymorphisms in populations (Nei 2005). It is worth noting that natural selection does not select for health but only for successful reproduction; thus, a gene will be retained even if it is detrimental to the health of the individual, as long as it does not reduce the number of surviving offspring. Therefore, whether the ALDH2 Glu504Lys allele is retained depends on whether the mutation affects the number of surviving offspring under certain external environmental conditions. Alcohol consumption behavior is one of the environmental factors most closely associated with ALDH2. The ALDH2 Glu504Lys allele not only causes individuals to experience adverse reactions such as blushing after alcohol consumption, but it also increases the risk of heart disease (Shen et al. 1863), esophageal cancer (Suo et al. 2019), gastric cancer (Hidaka et al. 2015), liver cancer (Sakamoto et al. 2006), and other diseases in alcohol drinkers. Nevertheless, during the evolution of the ALDH2 gene, alcohol consumption did not eliminate the deleterious mutation, i.e., Glu504Lys, by negative selection or even reduce its frequency. Although the exact reason for this remains unknown, it may be because wine originated after this mutation and the ALDH2 Glu504Lys allele was stable in the population before humans learned to produce wine. In addition, as large-scale brewing was available in ancient times, alcohol consumption was low, and the amount produced was small. Therefore, the disadvantages of the mutated gene were not obvious in such an environment of a relatively low frequency of alcohol consumption and low amounts. Furthermore, the ALDH2 Glu504Lys allele can even prevent individuals from drinking due to the “blushing syndrome” and other adverse reactions. As we know, some genes that were beneficial or at least harmless in the ancient environment can cause problems in the modern world. Just as the “thrifty genes” would be advantageous under the conditions of unpredictably alternating feast and famine that characterized the traditional human lifestyle, but they would lead to obesity and diabetes in the modern world (Diamond 2003). In conclusion, the effect of alcohol consumption on individuals carrying the ALDH2 Glu504Lys allele would not likely affect the number of surviving offspring or eliminate them through selection, at least in the ancient environment. It has been suggested that hepatitis B virus infection plays a selective role in this process, with the ALDH2 Glu504Lys allele being evolutionarily advantageous for hepatitis B virus carriers (Lin and Cheng 2002). In terms of geographical distribution, the infection rate of hepatitis B virus in East Asia is significantly higher than that in Europe and America (Schweitzer et al. 2015); in China, the rate is higher in the southeast coastal region and lower in the western region (Beasley et al. 1982). Hence, there may be a geographical correlation between HBV prevalence and ALDH2 deficiency within China and some other places to some degree. Studies have shown that the ALDH2 Glu504Lys allele increases the risk of liver cancer in alcohol drinkers (Sakamoto et al. 2006; Abe et al. 2015), but no increased risk is observed in non-drinkers (Liu et al. 2016). As both alcohol consumption and hepatitis B virus infection are the risk factors for liver cancer, those carrying the ALDH2 Glu504Lys allele may reduce alcohol consumption among hepatitis B virus-infected individuals, indirectly reducing their risk of develo** liver cancer (Liu et al. 2016). In addition, the ALDH2 Glu504Lys allele may attenuate liver damage in alcohol drinkers, with reduced AST, ALT, and GGT levels observed in these individuals (Takeshita et al. 2000). The mechanism may be explained by weakened acetaldehyde metabolism, reduced NADH production and oxidative stress (Matsumoto et al. 2007), as well as inhibition of NF-kB and MAPK pathways, decreased TNF-a production, and attenuated hepatic inflammatory response by increased ethanol load (Matsumoto et al. 2008; Lindros et al. 1999). Thus, on the one hand, the ALDH2 Glu504Lys allele alleviates liver damage in hepatitis B virus-infected individuals; on the other hand, it reduces the risk of liver cancer by reducing alcohol consumption, thus conferring these individuals with a better chance of survival. Hepatitis B virus infection may also exert a positive selection effect on the ALDH2 Glu504Lys allele, favoring its retention. In particular, individuals heterozygous for ALDH2 Glu504Lys may exhibit a heterozygote advantage, experiencing both a hepatoprotective effect not found in those homozygous for the wildtype gene and higher enzyme activity than in those homozygous for the mutant, which may allow the allele to reach a high frequency in some populations. A classic case is that the high frequency of the sickle cell haemoglobin (HbS) gene in malaria-endemic regions is thought to be related to a heterozygote (HbAS) advantage against fatal malaria. It has been shown that heterozygotes (HbAS) can provide significant protection against all-cause mortality and severe malarial anaemia compared with HbAA genotypes (Aidoo et al. 2002). Another possible selection factor is related to oxygen concentrations. ALDH2 has been shown to be essential for tolerance to hypoxic conditions, and it plays a protective role in hypoxia-related diseases such as ischemic heart disease (Liu et al. 2015; Sun et al. 2014) and hypoxic pulmonary hypertension (Zhao et al. 2019). Although the ALDH2 Glu504Lys allele has a high frequency in East Asia in general, it is less frequently distributed in the Tibetan population (4.4%) than in the Han populations of Guangdong (24.8%), Qingdao (17.6%), and Liaoning (12.2%) (Luo et al. 2009), and this difference may be related to the low oxygen environment of the plateau in Tibet. After the divergence of the common ancestors of the Han and Tibet populations in the early Neolithic period, the Han people migrated eastward into the plains, whereas Tibetans settled westward in the plateau region (Shi et al. 2005). Negative selection of the ALDH2 Glu504Lys allele under the low oxygen environment may have occurred in this population entering Tibet, resulting in its reduced frequency. In contrast, the ALDH2 Glu504Lys allele maintained a higher frequency in the Han population living in the plains, especially in the densely vegetated and humid southeastern coastal zone, because its disadvantage was not revealed due to sufficient oxygen in the environment. In addition, a higher rate of EGLN1 gene mutation has been demonstrated in Tibetans, reducing EGLN1 activity and facilitating adaptation to hypoxic conditions (Simonson et al. 2010). It was shown that ALDH2 activity in isolated hepatocytes of EGLN1-deficient mice was increased by 25% compared to wildtype and that levels of ethanol metabolism and ROS (reactive oxygen species) clearance were increased (Laitakari et al. 2019). It has been suggested that the hypoxic environment may also indirectly affect the activity of ALDH2 through a selective effect on the EGLN1 gene. The population in the plains is lack of EGLN1 gene mutation, and if they carry the ALDH2 Glu504Lys allele, it will be more difficult for them to adapt to the hypoxic environment in the Tibetan plateau. However, studies to date have not resolved this issue, and the relationship remains to be proven by further evidence. In conclusion, it is difficult to obtain a clear explanation for the evolutionary mechanism of the ALDH2 gene based on the findings thus far, and multiple factors may have played a role in the selection of the ALDH2 Glu504Lys allele. Ohta proposed the near-neutrality theory of evolution, in which mutation, genetic drift, and selection act simultaneously in evolution (Ohta 2002), which may be able to explain the evolution of ALDH2. A random mutation produced the ALDH2 Glu504Lys allele, genetic drift altered the gene frequency of this gene in the initial small population, and natural selection caused its differental distribution in different environments. Since the Neolithic period, populations around the world have been undergoing continuous migration and gene exchange (Yang and Fu 2018), and the spread of the ALDH2 Glu504Lys allele may also be associated with the migration of East Asian populations. Studies have shown that evolution in East Asia has been continuous since Homo erectus, and no large-scale replacement of native populations by foreign populations has occurred during this time. Although a small amount of integration of foreign populations with native subject populations occurred, this exchange of genes is minor compared to the intergenerational transmission of native populations (Yang and Fu 2018). This would explain why the spread of the ALDH2 Glu504Lys allele was confined to East Asia, without contributing to the human gene pool outside Asia. Two-way interaction and integration of northern and southern populations in East Asia began in the early Neolithic, and three southward migrations occurred, allowing the spread of East Asian populations to Southeast Asia and the Southwest Pacific islands (Zhang and Fu 2020). The ALDH2 Glu504Lys allele present in Thai and Cambodian populations (Goedde et al. 1992; Oota et al. 2004) may be derived from these migrations; the ALDH2 Glu504Lys allele also has a high frequency in South Korea and Japan (Goedde et al. 1992; Luo et al. 2009). Y-chromosome polymorphism suggests that the Japanese population originated from at least two migrations from mainland Asia (Hammer et al. 2006), and the Korean population may have originated from Northern Asian settlement and range expansion mostly from southern-to-northern China (** et al. 2003). In conclusion, Neolithic and subsequent East Asian population migrations played an important role in the spread of ALDH2 Glu504Lys alleles. Although the exact correlation between the migrations of East Asian populations and the dispersal of ALDH2 Glu504Lys alleles have not been confirmed, the distribution of the ALDH2 Glu504Lys allele after this event is quite similar to that of the present day. Most diseases are caused by a complex combination of genetic and environmental factors, and the causative genetic factors are largely associated with the significant influence of environmental adaptations in human evolution. The ALDH2 gene, an important genetic factor, is not only associated with the risk of digestive tumors associated with alcohol consumption (Suo et al. 2019; Hidaka et al. 2015; Sakamoto et al. 2006) but also contributes to the prevention of alcohol abuse (Shen et al. 1863), ischemia (Liu et al. 2015; Sun et al. 2014), metabolic disorders (Wang et al. 2016), and other adverse factors causing cardiac dysfunction. Although the evolution of the ALDH2 gene has not been elucidated, environmental factors, one of the most important drivers of evolution, undoubtedly played an important role in this process. Therefore, studying the interaction of the ALDH2 gene with specific environments and contexts can help to achieve precision medicine based on genetic and environmental factors for specific populations. For patients with cardiovascular diseases, we should examine their ALDH2 genotype and living environment, whether they have consumed alcohol for a long time or live in high-elevation hypoxic conditions to achieve a more personalized and precise treatment plan. For individuals with ALDH2 mutations, several therapies have been shown to increase the activity of the enzyme (Li et al. 2020; Sun et al. 2017). Therefore, in the future treatment of cardiovascular and other diseases, we may expect to implement ALDH2-based precision medical strategies to improve the health status of people in East Asia and even the global populations.Origin of the ALDH2 Glu504Lys Allele
Selection of the ALDH2 Glu504Lys Allele
Relationship with Alcohol Consumption
Relationship with Hepatitis B Virus Infection
Relationship with High-Elevation Hypoxic Environments
Dispersal of the ALDH2 Glu504Lys Allele
Perspective
Availability of data and material
Not applicable.
Code availability
Not applicable.
References
Abe H, Aida Y, Seki N, Sugita T, Tomita Y, Nagano T, Itagaki M, Sutoh S, Nagatsuma K, Itoh K, Matsuura T, Aizawa Y (2015) Aldehyde dehydrogenase 2 polymorphism for development to hepatocellular carcinoma in East Asian alcoholic liver cirrhosis. J Gastroenterol Hepatol 30(9):1376–1383. https://doi.org/10.1111/jgh.12948
Aidoo M, Terlouw DJ, Kolczak MS, McElroy PD, ter Kuile FO, Kariuki S, Nahlen BL, Lal AA, Udhayakumar V (2002) Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet 359(9314):1311–1312. https://doi.org/10.1016/S0140-6736(02)08273-9
Beasley RP, Lin CC, Chien CS, Chen CJ, Hwang LY (1982) Geographic distribution of HBsAg carriers in China. Hepatology 2(5):553–556. https://doi.org/10.1002/hep.1840020507
Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, de Ferranti SD, Ferguson JF, Fornage M, Gillespie C, Isasi CR, Jiménez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Lutsey PL, Mackey JS, Matchar DB, Matsushita K, Mussolino ME, Nasir K, O'Flaherty M, Palaniappan LP, Pandey A, Pandey DK, Reeves MJ, Ritchey MD, Rodriguez CJ, Roth GA, Rosamond WD, Sampson UKA, Satou GM, Shah SH, Spartano NL, Tirschwell DL, Tsao CW, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P (2018) American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee. Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association. Circulation 137(12):e67–e492. https://doi.org/10.1161/CIR.0000000000000558. Accessed Mar 20 2018
Diamond J (2003) The double puzzle of diabetes. Nature 423(6940):599–602. https://doi.org/10.1038/423599a
Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckmann G, Bhatia K, Chen LZ, Fang B, Lisker R et al (1992) Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet 88(3):344–346. https://doi.org/10.1007/BF00197271
Goldman D, Enoch MA (1990) Genetic epidemiology of ethanol metabolic enzymes: a role for selection. World Rev Nutr Diet 63:143–160. https://doi.org/10.1159/000418505
Hammer MF, Karafet TM, Park H, Omoto K, Harihara S, Stoneking M, Horai S (2006) Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes. J Hum Genet 51(1):47–58. https://doi.org/10.1007/s10038-005-0322-0
Hidaka A, Sasazuki S, Matsuo K, Ito H, Sawada N, Shimazu T, Yamaji T, Iwasaki M, Inoue M, Tsugane S, Group JS (2015) Genetic polymorphisms of ADH1B, ADH1C and ALDH2, alcohol consumption, and the risk of gastric cancer: the Japan Public Health Center-based prospective study. Carcinogenesis 36(2):223–231. https://doi.org/10.1093/carcin/bgu244
Ingman M, Kaessmann H, Paabo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408(6813):708–713. https://doi.org/10.1038/35047064
Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V (2011) Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics 5(4):283–303. https://doi.org/10.1186/1479-7364-5-4-283
** HJ, Kwak KD, Hammer MF, Nakahori Y, Shinka T, Lee JW, ** F, Jia X, Tyler-Smith C, Kim W (2003) Y-chromosomal DNA haplogroups and their implications for the dual origins of the Koreans. Hum Genet 114(1):27–35. https://doi.org/10.1007/s00439-003-1019-0
Laitakari A, Ollonen T, Kietzmann T, Walkinshaw G, Mennerich D, Izzi V, Haapasaari KM, Myllyharju J, Serpi R, Dimova EY, Koivunen P (2019) Systemic inactivation of hypoxia-inducible factor prolyl 4-hydroxylase 2 in mice protects from alcohol-induced fatty liver disease. Redox Biol 22:101145. https://doi.org/10.1016/j.redox.2019.101145
Li Y, Zhang D, ** W, Shao C, Yan P, Xu C, Sheng H, Liu Y, Yu J, **e Y, Zhao Y, Lu D, Nebert DW, Harrison DC, Huang W, ** L (2006) Mitochondrial aldehyde dehydrogenase-2 (ALDH2) Glu504Lys polymorphism contributes to the variation in efficacy of sublingual nitroglycerin. J Clin Invest 116(2):506–511. https://doi.org/10.1172/JCI26564
Li H, Borinskaya S, Yoshimura K, Kal’ina N, Marusin A, Stepanov VA, Qin Z, Khaliq S, Lee MY, Yang Y, Mohyuddin A, Gurwitz D, Mehdi SQ, Rogaev E, ** L, Yankovsky NK, Kidd JR, Kidd KK (2009) Refined geographic distribution of the oriental ALDH2*504Lys (nee 487Lys) variant. Ann Hum Genet 73(Pt 3):335–345. https://doi.org/10.1111/j.1469-1809.2009.00517.x
Li W, Shen C, Yin L, Ge J, Sun A (2019) Aldehyde Dehydrogenase 2 and heart failure. Adv Exp Med Biol 1193:89–106. https://doi.org/10.1007/978-981-13-6260-6_5
Li W, Yin L, Sun X, Wu J, Dong Z, Hu K, Sun A, Ge J (2020) Alpha-lipoic acid protects against pressure overload-induced heart failure via ALDH2-dependent Nrf1-FUNDC1 signaling. Cell Death Dis 11(7):599. https://doi.org/10.1038/s41419-020-02805-2
Lin YP, Cheng TJ (2002) Why can’t Chinese Han drink alcohol? Hepatitis B virus infection and the evolution of acetaldehyde dehydrogenase deficiency. Med Hypotheses 59(2):204–207. https://doi.org/10.1016/s0306-9877(02)00253-0
Lindros KO, Jokelainen K, Nanji AA (1999) Acetaldehyde prevents nuclear factor-kappa B activation and hepatic inflammation in ethanol-fed rats. Lab Invest 79(7):799–806
Liu X, Sun X, Liao H, Dong Z, Zhao J, Zhu H, Wang P, Shen L, Xu L, Ma X, Shen C, Fan F, Wang C, Hu K, Zou Y, Ge J, Ren J, Sun A (2015) Mitochondrial Aldehyde Dehydrogenase 2 regulates revascularization in chronic ischemia: potential impact on the development of coronary collateral circulation. Arterioscler Thromb Vasc Biol 35(10):2196–2206. https://doi.org/10.1161/ATVBAHA.115.306012
Liu J, Yang HI, Lee MH, Jen CL, Hu HH, Lu SN, Wang LY, You SL, Huang YT, Chen CJ (2016) Alcohol drinking mediates the association between Polymorphisms of ADH1B and ALDH2 and Hepatitis B-related Hepatocellular Carcinoma. Cancer Epidemiol Biomarkers Prev 25(4):693–699. https://doi.org/10.1158/1055-9965.EPI-15-0961
Luo HR, Wu GS, Pakstis AJ, Tong L, Oota H, Kidd KK, Zhang YP (2009) Origin and dispersal of atypical aldehyde dehydrogenase ALDH2487Lys. Gene 435(1–2):96–103. https://doi.org/10.1016/j.gene.2008.12.021
Lynch M, Ackerman MS, Gout JF, Long H, Sung W, Thomas WK, Foster PL (2016) Genetic drift, selection and the evolution of the mutation rate. Nat Rev Genet 17(11):704–714. https://doi.org/10.1038/nrg.2016.104
Masel J (2011) Genetic drift. Curr Biol 21(20):R837-838. https://doi.org/10.1016/j.cub.2011.08.007
Matsumoto A, Ichiba M, Horita M, Yamashita Z, Takahashi T, Isse T, Oyama T, Kawamoto T, Tomokuni K (2007) Lack of aldehyde dehydrogenase ameliorates oxidative stress induced by single-dose ethanol administration in mouse liver. Alcohol 41(1):57–59. https://doi.org/10.1016/j.alcohol.2007.01.004
Matsumoto A, Kawamoto T, Mutoh F, Isse T, Oyama T, Kitagawa K, Nakayama KI, Ichiba M (2008) Effects of 5-week ethanol feeding on the liver of aldehyde dehydrogenase 2 knockout mice. Pharmacogenet Genomics 18(10):847–852. https://doi.org/10.1097/FPC.0b013e328307a0a9
Matsumura Y, Stiles KM, Reid J, Frenk EZ, Cronin S, Pagovich OE, Crystal RG (2019) Gene therapy correction of Aldehyde Dehydrogenase 2 deficiency. Mol Ther Methods Clin Dev 15:72–82. https://doi.org/10.1016/j.omtm.2019.08.004
Nei M (2005) Selectionism and neutralism in molecular evolution. Mol Biol Evol 22(12):2318–2342. https://doi.org/10.1093/molbev/msi242
Ohta T (2002) Near-neutrality in evolution of genes and gene regulation. Proc Natl Acad Sci U S A 99(25):16134–16137. https://doi.org/10.1073/pnas.252626899
Oota H, Pakstis AJ, Bonne-Tamir B, Goldman D, Grigorenko E, Kajuna SL, Karoma NJ, Kungulilo S, Lu RB, Odunsi K, Okonofua F, Zhukova OV, Kidd JR, Kidd KK (2004) The evolution and population genetics of the ALDH2 locus: random genetic drift, selection, and low levels of recombination. Ann Hum Genet 68(Pt 2):93–109. https://doi.org/10.1046/j.1529-8817.2003.00060.x
Palanichamy MG, Sun C, Agrawal S, Bandelt HJ, Kong QP, Khan F, Wang CY, Chaudhuri TK, Palla V, Zhang YP (2004) Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet 75(6):966–978. https://doi.org/10.1086/425871
Sakamoto T, Hara M, Higaki Y, Ichiba M, Horita M, Mizuta T, Eguchi Y, Yasutake T, Ozaki I, Yamamoto K, Onohara S, Kawazoe S, Shigematsu H, Koizumi S, Tanaka K (2006) Influence of alcohol consumption and gene polymorphisms of ADH2 and ALDH2 on hepatocellular carcinoma in a Japanese population. Int J Cancer 118(6):1501–1507. https://doi.org/10.1002/ijc.21505
Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ (2015) Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 386(10003):1546–1555. https://doi.org/10.1016/S0140-6736(15)61412-X
Shaw LJ, Shaw RE, Merz CN, Brindis RG, Klein LW, Nallamothu B, Douglas PS, Krone RJ, McKay CR, Block PC, Hewitt K, Weintraub WS, Peterson ED, College A, of Cardiology-National Cardiovascular Data Registry I, (2008) Impact of ethnicity and gender differences on angiographic coronary artery disease prevalence and in-hospital mortality in the American College of Cardiology-National Cardiovascular Data Registry. Circulation 117(14):1787–1801. https://doi.org/10.1161/CIRCULATIONAHA.107.726562
Shen C, Wang C, Han S, Wang Z, Dong Z, Zhao X, Wang P, Zhu H, Sun X, Ma X, Zhu H, Zou Y, Hu K, Ge J, Sun A (2017) Aldehyde dehydrogenase 2 deficiency negates chronic low-to-moderate alcohol consumption-induced cardioprotecion possibly via ROS-dependent apoptosis and RIP1/RIP3/MLKL-mediated necroptosis. Biochim Biophys Acta Mol Basis Dis 1863(8):1912–1918. https://doi.org/10.1016/j.bbadis.2016.11.016
Shi H, Dong YL, Wen B, **ao CJ, Underhill PA, Shen PD, Chakraborty R, ** L, Su B (2005) Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3–M122. Am J Hum Genet 77(3):408–419. https://doi.org/10.1086/444436
Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, **ng J, Jorde LB, Prchal JT, Ge R (2010) Genetic evidence for high-altitude adaptation in Tibet. Science 329(5987):72–75. https://doi.org/10.1126/science.1189406
Sun A, Ren J (2013) ALDH2, a novel protector against stroke? Cell Res 23(7):874–875. https://doi.org/10.1038/cr.2013.76
Sun A, Zou Y, Wang P, Xu D, Gong H, Wang S, Qin Y, Zhang P, Chen Y, Harada M, Isse T, Kawamoto T, Fan H, Yang P, Akazawa H, Nagai T, Takano H, ** P, Komuro I, Ge J (2014) Mitochondrial aldehyde dehydrogenase 2 plays protective roles in heart failure after myocardial infarction via suppression of the cytosolic JNK/p53 pathway in mice. J Am Heart Assoc 3(5):e000779. https://doi.org/10.1161/JAHA.113.000779
Sun X, Zhu H, Dong Z, Liu X, Ma X, Han S, Lu F, Wang P, Qian S, Wang C, Shen C, Zhao X, Zou Y, Ge J, Sun A (2017) Mitochondrial aldehyde dehydrogenase-2 deficiency compromises therapeutic effect of ALDH bright cell on peripheral ischemia. Redox Biol 13:196–206. https://doi.org/10.1016/j.redox.2017.05.018
Suo C, Yang Y, Yuan Z, Zhang T, Yang X, Qing T, Gao P, Shi L, Fan M, Cheng H, Lu M, ** L, Chen X, Ye W (2019) Alcohol intake interacts with functional genetic Polymorphisms of Aldehyde Dehydrogenase (ALDH2) and Alcohol Dehydrogenase (ADH) to increase esophageal squamous cell cancer risk. J Thorac Oncol 14(4):712–725. https://doi.org/10.1016/j.jtho.2018.12.023
Takeshita T, Yang X, Morimoto K (2000) The ALDH2 genotype, alcohol intake, and liver-function biomarkers among Japanese male workers. Hum Genet 106(6):589–593. https://doi.org/10.1007/s004390000314
Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65(Pt 1):43–62. https://doi.org/10.1046/j.1469-1809.2001.6510043.x
Vasiliou V, Bairoch A, Tipton KF, Nebert DW (1999) Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal map**. Pharmacogenetics 9(4):421–434
Wang C, Fan F, Cao Q, Shen C, Zhu H, Wang P, Zhao X, Sun X, Dong Z, Ma X, Liu X, Han S, Wu C, Zou Y, Hu K, Ge J, Sun A (2016) Mitochondrial aldehyde dehydrogenase 2 deficiency aggravates energy metabolism disturbance and diastolic dysfunction in diabetic mice. J Mol Med (berl) 94(11):1229–1240. https://doi.org/10.1007/s00109-016-1449-5
Xu L, Zhao G, Wang J, Shen C, Li X, Lu F, Jiang H, Liu G, Hu K, Tang Y, Sun A, Ge J (2018) Impact of genetic variation in Aldehyde Dehydrogenase 2 and alcohol consumption on coronary artery lesions in Chinese patients with stable coronary artery disease. Int Heart J 59(4):689–694. https://doi.org/10.1536/ihj.17-165
Yang MA, Fu Q (2018) Insights into modern human prehistory using ancient genomes. Trends Genet 34(3):184–196. https://doi.org/10.1016/j.tig.2017.11.008
Zhang M, Fu Q (2020) Human evolutionary history in Eastern Eurasia using insights from ancient DNA. Curr Opin Genet Dev 62:78–84. https://doi.org/10.1016/j.gde.2020.06.009
Zhao Y, Wang B, Zhang J, He D, Zhang Q, Pan C, Yuan Q, Shi Y, Tang H, Xu F, Wei S, Chen Y (2019) ALDH2 (Aldehyde Dehydrogenase 2) protects against Hypoxia-induced pulmonary hypertension. Arterioscler Thromb Vasc Biol 39(11):2303–2319. https://doi.org/10.1161/ATVBAHA.119.312946
Zhou M, Wang H, Zeng X, Yin P, Zhu J, Chen W, Li X, Wang L, Wang L, Liu Y, Liu J, Zhang M, Qi J, Yu S, Afshin A, Gakidou E, Glenn S, Krish VS, Miller-Petrie MK, Mountjoy-Venning WC, Mullany EC, Redford SB, Liu H, Naghavi M, Hay SI, Wang L, Murray CJL, Liang X (2019) Mortality, morbidity, and risk factors in China and its provinces, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 394(10204):1145–1158. https://doi.org/10.1016/S0140-6736(19)30427-1
Zlotogora J (1994) High frequencies of human genetic diseases: founder effect with genetic drift or selection? Am J Med Genet 49(1):10–13. https://doi.org/10.1002/ajmg.1320490104
Funding
This work was supported by National Science Fund for Distinguished Young Scholars (81725002).
Author information
Authors and Affiliations
Contributions
XZ contributed to literature searching and wrote the manuscript. JG and AS supervised the study and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, X., Sun, A. & Ge, J. Origin and Spread of the ALDH2 Glu504Lys Allele. Phenomics 1, 222–228 (2021). https://doi.org/10.1007/s43657-021-00017-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43657-021-00017-y