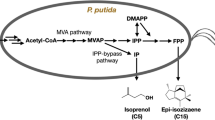
Abstract
Beyond its potential for biofuel production, Pseudomonas putida’s capability to metabolize lignin and other lignocellulosic materials earmarks it as a pivotal candidate for engineering to yield diverse value-added chemicals, thereby challenging traditional petrochemical approaches. Recognizing the inherent environmental, economic, and societal advantages, amplifying role of P. putida in industrial applications becomes imperative. In this context, our study focused on characterizing a comprehensive set of promoters and ribosome binding site tailored for P. putida, spanning a broad spectrum of activities. By leveraging these genetic tools, we adeptly balanced the heterologous mevalonate (MVA) pathway flux within P. putida. As a culmination of our efforts, the optimal MVA-producing strains were identified, achieving a remarkable yield of 5 g/LMVA in a 5 L fed-batch fermenter, marking the highest reported yield in Pseudomonas to date. This research not only provides valuable genetic tools for future engineering studies with P. putida, but also accentuates P. putida’s potential in synthetic biology and its promise for sustainable chemical production.




Similar content being viewed by others
Data availability
The data supporting the findings of this study are available upon request. Please contact the corresponding author for further details on accessing the data.
References
Wolf DE, Hoffman CH, Aldrich PE, et al. β-Hydroxy-β-methyl-δ-valerolactone (divalonic acid), a new biological factor. J Am Chem Soc. 1956;78:4499. https://doi.org/10.1021/ja01598a090.
Tamura G. Hiochic acid, a new growth factor for Lactobacillus homohiochi and Lactobacillus heterohiochi. J General Appl Microbiol. 2004;50(6):327–30. https://doi.org/10.2323/jgam.2.431.
Beck ZQ, Eliot AC, Peres CM, et al. Utilization of phosphoketolase in the production of mevalonate, isoprenoid precursors, and isoprene . Google Patents; 2015.
Zhang C, Schneiderman DK, Cai T, et al. Optically active β-methyl-δ-valerolactone: biosynthesis and polymerization. ACS Sustain Chem Eng. 2016;4(8):4396–402. https://doi.org/10.1021/acssuschemeng.6b00992.
**ong M, Schneiderman DK, Bates FS, et al. Scalable production of mechanically tunable block polymers from sugar. Proc Natl Acad Sci USA. 2014;111(23):8357–62. https://doi.org/10.1073/pnas.1404596111.
Liman GLS, Hulko T, Febvre HP, et al. A linear pathway for mevalonate production supports growth of Thermococcus kodakarensis. Extremophiles. 2019;23(2):229–38. https://doi.org/10.1007/s00792-019-01076-w.
Tsuruta H, Paddon C, Eng D, et al. High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS ONE. 2009;4:4489. https://doi.org/10.1371/journal.pone.0004489.
Tabata K, Hashimoto S. Production of mevalonate by a metabolically-engineered Escherichia coli. Biotechnol Lett. 2004;26(19):1487–91. https://doi.org/10.1023/B:BILE.0000044449.08268.7d.
Martin VJ, Pitera DJ, Withers ST, et al. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21(7):796–802. https://doi.org/10.1038/nbt833.
Pitera DJ, Paddon CJ, Newman JD, et al. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng. 2007;9(2):193–207. https://doi.org/10.1016/j.ymben.2006.11.002.
Satowa D, Fujiwara R, Uchio S, et al. Metabolic engineering of E. coli for improving mevalonate production to promote NADPH regeneration and enhance acetyl-CoA supply. Biotechnol Bioeng. 2020;117(7):2153–64. https://doi.org/10.1002/bit.27350.
Wang J, Niyompanich S, Tai YS, et al. Engineering of a highly efficient Escherichia coli strain for mevalonate fermentation through chromosomal integration. Appl Environ Microbiol. 2016;82(24):7176–84. https://doi.org/10.1128/aem.02178-16.
Wang Q, Xu J, Sun Z, et al. Engineering an in vivo EP-bifido pathway in Escherichia coli for high-yield acetyl-CoA generation with low CO2 emission. Metab Eng. 2019;51:79–87. https://doi.org/10.1016/j.ymben.2018.08.003.
Rodriguez S, Denby CM, Van Vu T, et al. ATP citrate lyase mediated cytosolic acetyl-CoA biosynthesis increases mevalonate production in Saccharomyces cerevisiae. Microbial Cell Fact. 2016;15(1):48. https://doi.org/10.1186/s12934-016-0447-1.
Bitzenhofer NL, Kruse L, Thies S, et al. Towards robust Pseudomonas cell factories to harbour novel biosynthetic pathways. Essays Biochem. 2021;65(2):319–36. https://doi.org/10.1042/ebc20200173.
Yang J, Im Y, Kim TH, et al. Engineering Pseudomonas putida KT2440 to convert 2,3-butanediol to mevalonate [1879-0909 (Electronic)].
Moser S, Pichler H. Identifying and engineering the ideal microbial terpenoid production host. Appl Microbiol Biotechnol. 2019;103(14):5501–16. https://doi.org/10.1007/s00253-019-09892-y.
Loeschcke A, Thies S. Pseudomonas putida—a versatile host for the production of natural products. Appl Microbiol Biotechnol. 2015;99(15):6197–214. https://doi.org/10.1007/s00253-015-6745-4.
Hernandez-Arranz S, Perez-Gil J, Marshall-Sabey D, et al. Engineering Pseudomonas putida for isoprenoid production by manipulating endogenous and shunt pathways supplying precursors. Microbial Cell Fact. 2019;18(1):152. https://doi.org/10.1186/s12934-019-1204-z.
Shcherbo D, Murphy CS, Ermakova GV, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418(3):567–74. https://doi.org/10.1042/bj20081949.
Duan Y, Zhang X, Zhai W, et al. Deciphering the rules of ribosome binding site differentiation in context dependence. ACS Synth Biol. 2022;11(8):2726–40. https://doi.org/10.1021/acssynbio.2c00139.
Lu S, Zhou C, Guo X, et al. Enhancing fluxes through the mevalonate pathway in Saccharomyces cerevisiae by engineering the HMGR and β-alanine metabolism. Microb Biotechnol. 2022;15(8):2292–306. https://doi.org/10.1111/1751-7915.14072.
Dudley QM, Anderson KC, Jewett MC. Cell-free mixing of Escherichia coli crude extracts to prototype and rationally engineer high-titer mevalonate synthesis. ACS Synth Biol. 2016;5(12):1578–88. https://doi.org/10.1021/acssynbio.6b00154.
Dueber JE, Wu GC, Malmirchegini GR, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27(8):753–9. https://doi.org/10.1038/nbt.1557.
Liu H, Chen Y, Zhang Y, et al. Enhanced production of polyhydroxyalkanoates in Pseudomonas putida KT2440 by a combination of genome streamlining and promoter engineering. Int J Biol Macromol. 2022;209(Pt A):117–24. https://doi.org/10.1016/j.ijbiomac.2022.04.004.
Zhang Y, Liu H, Liu Y, et al. A promoter engineering-based strategy enhances polyhydroxyalkanoate production in Pseudomonas putida KT2440. Int J Biol Macromol. 2021;191:608–17. https://doi.org/10.1016/j.ijbiomac.2021.09.142.
Wirth NT, Nikel PI. Combinatorial pathway balancing provides biosynthetic access to 2-fluoro-cis, cis-muconate in engineered Pseudomonas putida. Chem Catal. 2021;1(6):1234–59. https://doi.org/10.1016/j.checat.2021.09.002.
Pfleger BF, Pitera DJ, Smolke CD, et al. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24(8):1027–32. https://doi.org/10.1038/nbt1226.
Zelcbuch L, Antonovsky N, Bar-Even A, et al. Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucl Acids Res. 2013;41(9):e98. https://doi.org/10.1093/nar/gkt151.
Xu L, Liu P, Dai Z, et al. Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants. Microb Cell Fact. 2021;20(1):148. https://doi.org/10.1186/s12934-021-01641-z.
Jeschek M, Gerngross D, Panke S. Combinatorial pathway optimization for streamlined metabolic engineering. Curr Opin Biotechnol. 2017;47:142–51. https://doi.org/10.1016/j.copbio.2017.06.014.
Jeschek M, Gerngross D, Panke S. Rationally reduced libraries for combinatorial pathway optimization minimizing experimental effort. Nat Commun. 2016;7:11163. https://doi.org/10.1038/ncomms11163.
Englund E, Liang F, Lindberg P. Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci Rep. 2016;6:36640. https://doi.org/10.1038/srep36640.
Song AA, Abdullah JO, Abdullah MP, et al. Overexpressing 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) in the lactococcal mevalonate pathway for heterologous plant sesquiterpene production. PLoS ONE. 2012;7(12):e52444. https://doi.org/10.1371/journal.pone.0052444.
Wang Y, **g F, Yu S, et al. Co-overexpression of the HMGR and FPS genes enhances artemisinin content in Artemisia annua L. J Med Plants Res. 2011;8:4.
Author information
Authors and Affiliations
Contributions
LZ: Writing-Original Draft, Conceptualization, Methodology. TF: Validation, Investigation. XZ: Project administration. YC: Supervision, Project administration.
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, L., Fan, TP., Cai, Y. et al. Production of mevalonate in Pseudomonas putida via tuning the expression of pathway gene. Syst Microbiol and Biomanuf 4, 1162–1173 (2024). https://doi.org/10.1007/s43393-023-00225-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-023-00225-9