Abstract
The CRISPR/Cas9 technology revolutionizes targeted gene knockout in diverse organisms including plants. However, screening edited alleles, particularly those with multiplex editing, from herbicide- or antibiotic-resistant transgenic plants and segregating out the Cas9 transgene represent two laborious processes. Current solutions to facilitate these processes rely on different selection markers. Here, by taking advantage of the opposite functions of a d-amino acid oxidase (DAO) in detoxifying d-serine and in metabolizing non-toxic d-valine to a cytotoxic product, we develop a DAO-based selection system that simultaneously enables the enrichment of multigene edited alleles and elimination of Cas9-containing progeny in Arabidopsis thaliana. Among five DAOs tested in Escherichia coli, the one encoded by Trigonopsis variabilis (TvDAO) could confer slightly stronger d-serine resistance than other homologs. Transgenic expression of TvDAO in Arabidopsis allowed a clear distinction between transgenic and non-transgenic plants in both d-serine-conditioned positive selection and d-valine-conditioned negative selection. As a proof of concept, we combined CRISPR-induced single-strand annealing repair of a dead TvDAO with d-serine-based positive selection to help identify transgenic plants with multiplex editing, where d-serine-resistant plants exhibited considerably higher co-editing frequencies at three endogenous target genes than those selected by hygromycin. Subsequently, d-valine-based negative selection successfully removed Cas9 and TvDAO transgenes from the survival offspring carrying inherited mutations. Collectively, this work provides a novel strategy to ease CRISPR mutant identification and Cas9 transgene elimination using a single selection marker, which promises more efficient and simplified multiplex CRISPR editing in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
CRISPR/Cas9 is a revolutionary genome editing technology inspired by the adaptive immune system of bacteria and archaea for defending against viral infection and has been widely applied to gene knockout in various organisms including plants (Gürel et al. 2020). Current CRISPR/Cas9 applications in plants predominantly rely on Agrobacterium tumefaciens-mediated genetic transformation, by which the T-DNA encoding genome editing reagents are stably integrated into the plant genome. In the model species Arabidopsis thaliana, Cas9 overexpression by a strong constitutive promoter (e.g., 35S promoter) often results in chimeric mutations in the T1 generation (Feng et al. 2014; Tsutsui and Higashiyama 2017; Wolabu et al. 2020). In contrast, using an egg cell-specific promoter to drive Cas9 expression can produce nonchimeric (that is, heterozygous, homozygous or biallelic) T1 mutants, although with a relatively lower efficiency (Wang et al. 2023), base editing (Xu et al. 2020a), and prime editing (Xu et al. 2020b). CRISPR-mediated disruption of MAR1, an endogenous transporter gene, has been demonstrated to facilitate co-selection of target gene editing in Arabidopsis and tomato (Rinne et al. 2021). A visible selection system has also been established in Arabidopsis to enrich CRISPR-induced mutants in the T1 generation, which is based on a glabrous phenotype caused by mutating the GLABRA2 gene that is involved in trichome formation (Kong et al. 2021).
On the other hand, the retention of Cas9 transgene after target gene knockout can significantly increase the risk of off-target editing (Fu et al. 2013; Zhang et al. 2018; Cullot et al. 2019). Moreover, in transgenic complementation experiments for validating the mutant phenotype and target gene correlation, it is necessary to eliminate the Cas9 before the target gene is re-introduced into the edited mutant plant. Furthermore, in CRISPR-mediated molecular breeding, the inclusion of the Cas9 transgene in crops raises regulatory concerns. Although the Cas9 transgene can be eliminated through genetic segregation, the process of screening non-transgenic progeny with desired mutations is laborious. Therefore, several strategies have been developed to facilitate the isolation of Cas9-free mutant plants (He and Zhao 2019). One approach for eliminating Cas9 from genome-edited plants involves RNA interference-induced herbicide susceptibility, which allows the survival of Cas9-free plants (Lu et al. 2017). Alternatively, non-transgenic seeds or plants could be visually distinguished from transgenic ones by integrating a transgene giving rise to detectable fluorescence (Gao et al. 2016; Wang and Chen 2020), early flowering (Liu et al. 2019; Lao et al. 2023), or anthocyanin accumulation (Liu et al. 2019). Although He and colleagues have created an ingenious strategy, called TKC (Transgene Killer CRISPR), enabling self-elimination of the CRISPR construct after targeted gene editing in rice (He et al. 2018), this approach cannot be applied to Arabidopsis that is transformed by the floral dip method (Clough and Bent 1998) rather than tissue culture. Of note, to date, no strategy has been developed to enable both the enrichment of CRISPR-edited mutants and subsequent elimination of the Cas9 transgene from the mutant offspring.
d-amino acid oxidases (DAOs) are specialized enzymes for d-amino acid metabolism, which are encoded by many eukaryotes but not plants (Pilone 2000). Interestingly, d-serine at high concentrations is toxic to plants but can be detoxified by transgenically expressed DAOs (Erikson et al. 2004; Lim et al. 2007; Gisby et al. 2012). In contrast, d-valine is non-toxic to plants but can be metabolized to a toxic product by a DAO transgene (Erikson et al. 2004; Gisby et al. 2012). In this study, we leveraged the dual functions of a DAO in metabolizing different d-amino acid substrates to develop a surrogate selection system for simplifying genome editing in Arabidopsis. This system can simultaneously facilitate the screening of CRISPR-edited alleles and subsequent elimination of mutant progeny containing the Cas9 transgene.
Results
TvDAO confers d-serine resistance in Escherichia coli
To utilize a DAO-based surrogate selection marker for CRISPR editing, we first sought to screen a DAO with robust catalytic activity in detoxifying d-serine. For this purpose, we selected five homologous DAOs (Pilone 2000; Pollegioni et al. 2007, 2018) from Rhodotorula gracilis (RgDAO), Homo sapiens (hDAO D31H), Fusarium verticillioides (FvDAO), Trigonopsis variabilis (TvDAO), and Mus musculus (MmDAO), respectively. These DAOs were codon-optimized (Supplementary Sequences) for optimal expression in Arabidopsis before gene synthesis. When using the Arabidopsis protoplast-based transient assay to evaluate the activity of these DAOs in detoxifying d-serine, we observed that Arabidopsis protoplasts were not sensitive to d-serine treatment. Considering that E. coli is a frequently used heterologous system for evaluating the function of a DAO in detoxifying d-amino acids (Pollegioni et al. 2007), we switched to E. coli to compare the activities of these DAOs. In three biological replicates, E. coli cells expressing TvDAO exhibited slightly better growth on the d-serine-containing medium than those expressing the other DAOs, particularly at a d-serine concentration of 60 mM (Fig. 1). Conversely, E. coli cells expressing the negative control CERK1, which encodes a receptor-like kinase from Arabidopsis, suffered obvious growth arrest in the presence of d-serine (Fig. 1). These findings suggest that all DAO transgenes are able to confer d-serine tolerance in E. coli.
d-amino acid oxidase confers d-serine tolerance in Escherichia coli. E. coli cells expressing Rhodotorula gracilis d-amino acid oxidase (RgDAO), Homo sapiens DAO (hDAO D31H), Fusarium verticillioides DAO (FvDAO), Trigonopsis variabilis DAO (TvDAO), or Mus musculus DAO (MmDAO) were diluted to indicated cell concentrations (OD600) and inoculated on the growth medium containing 0, 50 or 60 mM d-serine. E. coli cells expressing Arabidopsis thaliana CERK1 encoding a receptor kinase were used as a negative control. TvDAO used for further study is highlighted in red. These experiments were repeated three times with similar results
TvDAO enables both positive and negative selection for transgenic Arabidopsis plants
Considering that the TvDAO is not only smaller in size but also exhibited modestly stronger activity in conferring d-serine resistance in E. coli (Fig. 1), we next focused on TvDAO and attempted to validate whether it could be employed for d-serine-based positive selection and d-valine-based negative selection of transgenic plants. To this end, we generated transgenic Arabidopsis plants expressing TvDAO along with the HygR gene (Fig. 2A) for side-by-side comparison. When screening the transgenic T1 plants on 1/2 Murashige and Skoog (MS) medium containing 5 mM d-serine or 25 mg L−1 hygromycin, the transgenic plants selected by d-serine exhibited a clearer resistance phenotype than those selected by hygromycin (Fig. 2B), implying that TvDAO may be a better selection marker gene than HygR in Arabidopsis.
TvDAO enables both positive and negative selection of transgenic Arabidopsis plants. A Diagram of the expression cassettes for TvDAO and hygromycin resistance gene (HygR) in the binary plasmid. LB, T-DNA left border; RB, T-DNA right border; HSPter, Heat shock protein terminator. B Transgenic TvDAO plant selected by d-serine shows a clearer resistance phenotype than that selected by hygromycin. Seedlings indicated by white arrows are transgenic plants. Scale bar = 1 cm. C Progeny of d-serine-resistant lines, TvDAO #2 and TvDAO #7, show resistance segregation in both d-serine-based positive selection and d-valine-based negative selection. Scale bar = 1 cm. D PCR-based genoty** reveals that d-valine resistance or susceptibility is consistent with the absence or presence of TvDAO transgene. a1-a3 and d1-d3 are randomly selected alive or dying seedlings, respectively, on the d-valine-containing medium. ACTIN2 was used as an internal control
The T2 generation of two representative lines, TvDAO #2 and TvDAO #7, showed resistance segregation when germinated on the medium containing d-serine or d-valine, whereas all wild-type plants were sensitive to d-serine but grew well on the d-valine-containing medium (Fig. 2C). Consistently, PCR-based genoty** of three randomly selected healthy seedlings (i.e., a1-a3) or dying seedlings (i.e., d1-d3) from the progeny of TvDAO #2 or TvDAO #7 grown in the presence of d-valine showed that the a1-a3 were transgene-free, while the d1-d3 were transgenic (Fig. 2D). These results indicated that the TvDAO-based reporter supports both positive selection of transgenic plants by d-serine and negative selection of transgene-free plants by d-valine.
TvDAO-based selection system facilitates the screening of CRISPR-edited alleles in Arabidopsis
Given that TvDAO can be used for both positive and negative selection in Arabidopsis, depending on the d-amino acid substrate, we hypothesized that it would serve as an excellent surrogate selection marker for enriching CRISPR-edited alleles and then eliminating Cas9-containing progeny. To achieve this, CRISPR-induced single-strand annealing (SSA) repair converting a dead TvDAO (dTvDAO) reporter to an active form was devised (Fig. 3A, and B). SSA is a homologous recombination-based DNA repair mechanism that promotes recombination between two repetitive DNA sequences flanking a double-stranded break (DSB), resulting in the deletion of the intervening fragment (Endo et al. 2021; Vu et al. 2022). To validate the effectiveness of SSA repair in plant cells, we tested a dead GFP (dGFP) reporter (Fig. S1A), in which GFP was split into two fragments sharing a 321-bp overlap** sequence (nucleotides 199–519). These two fragments were separated by a linker containing two consecutive stop codons and a human genome-derived Cas9 target site (TGATGA-Cas9 TS). To optimize the CRISPR-induced SSA efficiency, we compared three different human target sites (i.e., hEMX1-1, hEMX1-2 and hEfemp1) in the dGFP reporter using corresponding gRNAs, namely hEMX1-gRNA1, hEMX1-gRNA2 (Cong et al. 2013), and hEfemp1-gRNA (Xu et al. 2020a). None of these gRNAs have predictable off-target sites in Arabidopsis. Transfection of the dGFP reporters alone into Arabidopsis protoplasts failed to produce any GFP fluorescence, whereas co-expressing Cas9 and a gRNA targeting the cognate human target site efficiently restored GFP expression (Fig. S1B–D). Considering comparable GFP regeneration efficiencies between the three human target site/gRNA pairs, we chose the hEfemp1 TS/hEfemp1-gRNA pair to build the dTvDAO reporter because this pair has been used in planta in a recent study (Xu et al. 2020a).
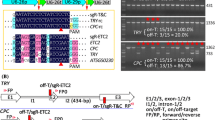
TvDAO helps to enrich CRISPR-edited alleles in Arabidopsis. A Diagram of the expression cassettes for dTvDAO, Cas9 (SpCas9), gRNAs and hygromycin resistance gene (HygR) in the binary plasmid. NLS, nuclear localization signal; E9ter, E9 terminator. B Rationale of CRISPR-induced single-strand annealing (SSA) repair to restore a functional TvDAO selection marker. In the dead TvDAO (dTvDAO) reporter, the most 5′ (1–279) and 3′ segments (789–1068) of TvDAO were colored in green. The middle segment (280–788) of TvDAO was colored in blue and the two copies (LH and RH) of it were spacered by two consecutive stop codons and a Cas9 target site from the human hEfemp1 locus. C PCR-based genoty** confirms successful SSA repair of dTvDAO in d-serine-resistant transgenic T1 plants. S, d-serine selection. H, hygromycin selection. Transgenic TvDAO #2 plant was used as a positive control. D Sanger sequencing of target amplicons validates the editing of all four target sites (dTvDAO, CPC, ETC2 and TRY) in the dTvDAO-gRNAs-Cas9 #S1 (Cas9 #S1) plant. Blue letters in dTvDAO represent the C-terminal coding sequences of LH and RH, while green letters represent the coding sequences adjacent to the C-terminus of RH. Black bold letters mark PAMs and target sequences of gRNAs are underlined. Deletions are indicated by red dashes, while insertions are colored in red. E The Cas9 #S1 plant exhibits visible clustering of trichomes on the leaf surface as indicated by red arrows. Scale bar = 1 cm. F Summary of mutation frequencies at individual target sites in transgenic T1 plants selected by d-serine or hygromycin. HO, HE, Bi and WT denote homozygote, heterozygote, bi-allele and wild type, respectively
We designed the dTvDAO reporter by duplicating nucleotides 280–788 of the TvDAO coding sequence and inserting the linker of TGATGA-hEfemp1 TS between the two duplicated sequences (i.e., LH and RH) (Fig. 3B). We constructed the binary plasmid dTvDAO-gRNAs-Cas9 [pHEE401E-UBQ10pro::dTvDAO-poly A-(tRNA- gRNA)s] to express Cas9 using the egg cell-specific EC1.2-EC1.1 fusion (EC1f) promoter (Wang et al. 3C and S2). This indicated successful recovery of TvDAO by SSA, which was further confirmed by Sanger sequencing of TvDAO amplicons from these plants (Fig. 3D). Among a total of 40 hygromycin-resistant plants obtained, 36 (that is, H1-H40 except H9, H14, H15 and H23) were identified as transgenic plants, which produced PCR amplicons of edited or non-edited dTvDAO (Figs. 3C and S2).
All 27 d-serine-resistant transgenic T1 plants harbored mutations for at least two of the three endogenous target genes (Fig. S3). Strikingly, 25 out of the 27 (92.6%) contained concurrent mutations across all three genes (Figs. 3D and S3). Consistently, slightly clustered trichomes could be seen on the leaf surface of some plants (Fig. 3E), as exemplified by Cas9 #S1 that harbored homozygous mutation in CPC and heterozygous mutations in ETC2 and TRY (Figs. 3D and S3). By contrast, only 19 out of the 36 (52.8%) hygromycin-resistant transgenic T1 plants contained concurrent mutations for all three target genes (Fig. S3). Notably, there were five plants (that is, H1, H27 and H33-35) containing no mutations at all for the four target loci including dTvDAO (Figs. S3 and S4). Importantly, d-serine-resistant transgenic plants not only showed an exceedingly higher co-editing frequency but also higher editing frequencies at individual endogenous target sites than hygromycin-resistant transgenic plants, namely 100% versus 66.7% for CPC, 96.3% versus 61.1% for ETC2, and 96.3% versus 77.8% for TRY (Fig. 3F). These findings demonstrated that the dTvDAO surrogate selection marker can ease the screening of mutant alleles produced by multiplex CRISPR editing in Arabidopsis.
TvDAO-based selection system facilitates the identification of Cas9-free progeny
Next, we evaluated whether the TvDAO-based selection system could also be used for identifying Cas9-free mutant progeny. To this end, we germinated the T2 progeny of the Cas9 #S1 plant on the 1/2 MS medium, with or without 15 mM d-valine. As anticipated, some seedlings were sensitive to d-valine (Fig. 4A). PCR-based genoty** revealed that three randomly selected healthy seedlings (i.e., A1-A3) were all Cas9- and TvDAO-free, whereas three randomly selected dying seedlings (i.e., D1-D3) all contained the transgenes (Fig. 4B). Sanger sequencing of target amplicons validated stable inheritance of T1 mutations at all three endogenous target sites in the T2 plant Cas9 #S1-A3, in which the mutations at the ETC2 and TRY loci became homozygous (Fig. 4C). Therefore, the Cas9 #S1-A3 plant represented a Cas9-free cpc etc2 try triple mutant allele, which displayed highly clustered trichomes (Fig. 4D) as previously reported (Kirik et al. 2004). These results suggested that d-valine-based negative selection allows efficient identification of Cas9-free mutant alleles.
TvDAO can be used to eliminate Cas9-carrying mutant progeny. A Progeny of the transgenic Cas9 #S1 plant exhibit resistance segregation on 1/2 MS medium containing 15 mM d-valine. Scale bar = 1 cm. B PCR-based genoty** confirms transgene elimination in survival T2 plants of Cas9 #S1 under d-valine-conditioned negative selection. A1-A3 and D1-D3 are randomly selected alive or dying seedlings, respectively. C Sanger sequencing of target amplicons validates inherited mutations at the three endogenous target sites in the T2 plant Cas9 #S1-A3. Black bold letters mark PAMs and target sequences of gRNAs are underlined. Insertion and deletions are in red. D The Cas9 #S1-A3 plant displays highly clustered trichomes as indicated by red arrows. Scale bar = 1 cm
Discussion
Screening Cas9-induced mutant alleles from herbicide- or antibiotic-resistant transgenic plants and isolating Cas9-free mutant progeny are two labor-consuming processes during plant genome editing. Although several surrogate selection systems have been explored to facilitate either process (Liu et al. 2019; Wang and Chen 2020; Xu et al. 2020a, 2020b; Kong et al. 2021; Rinne et al. 2021; Lao et al. 2015), resulting in the pHEE401E-UBQ10pro::dTvDAO-ploy A (dTvDAO-Cas9) plasmid. The specific gRNAs targeting the three Arabidopsis endogenous genes were designed using CRISPR-GE (**e et al. 2017). The polycistronic tRNA-gRNA repeats were synthesized through Golden Gate assembly, then amplified by PCR using primer pairs tRNA-F/HSP-R and inserted into the I-CeuI site of the dTvDAO-Cas9 plasmid to obtain the pHEE401E-UBQ10pro::dTvDAO-ploy A-(tRNA-gRNA)s (dTvDAO-gRNAs-Cas9) plasmid. The primers used for plasmid construction in this work are listed in Supplementary Table S1.
D-serine tolerance assay in E. coli
The E. coli cells expressing RgDAO, hDAO D31H, FvDAO, TvDAO, MmDAO, or CERK1 were cultured at 37 °C overnight. The cultures were then diluted to an OD600 of 0.5, 0.1, 0.02, and 0.005 before being applied onto LB medium containing 50 mg L−1 ampicillin, with or without 50 mM or 60 mM d-serine. The plates were incubated at 37 °C for approximately 20 h, and the tolerance phenotypes were recorded using a camera.
Protoplast isolation and transfection
Arabidopsis protoplasts were extracted and transfected as previously described, using the leaves of 4-week-old plants (Yoo et al. 2007). The dGFP reporter was transiently expressed alone or co-expressed with Cas9 and a cognate gRNA to the human target site in the dGFP reporter in Arabidopsis protoplasts. After overnight culture, the fluorescence was observed using a Leica DM8i C inverted microscope (Leica Biosystems, Germany).
Generation of transgenic plants
The binary plasmids TvDAO, dTvDAO-Cas9, or dTvDAO-gRNAs-Cas9 were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. The Agrobacterium cells carrying the appropriate binary plasmids were used to transform Arabidopsis using the widely used floral dip method (Clough and Bent 1998).
Genomic DNA extraction
Crude genomic DNA (gDNA) extracts of Arabidopsis plants were obtained by homogenizing three leaves from a single plant in TKE buffer (100 mM Tris-HCl, pH 9.5, 1 M KCl, 10 mM EDTA), followed by incubation at 70 °C for 30 min. The tenfold diluted gDNA extract was used as a PCR template for genoty**.
Genoty** of Arabidopsis plants
The Green Taq Mix (Vazyme, China) was used for genoty**. The PCR amplicons were separated by agarose gel electrophoresis. The gel strips containing PCR amplicons were excised and used for Sanger sequencing to validate the sequences. The primers used for genoty** PCR in this work are listed in Supplementary Table S1.
Data availability
All data generated in this study are available in the paper or online Supplementary Information.
References
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Cullot G, Boutin J, Toutain J, Prat F, Pennamen P, Rooryck C, Teichmann M, Rousseau E, Lamrissi-Garcia I, Guyonnet-Duperat V, Bibeyran A, Lalanne M, Prouzet-Mauléon V, Turcq B, Ged C, Blouin JM, Richard E, Dabernat S, Moreau-Gaudry F, Bedel A (2019) CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun 10:1136
de Jong OG, MurphyDE MI, Willms E, Garcia-Guerra A, Gitz-Francois JJ, Lefferts J, Gupta D, Steenbeek SC, van Rheenen J, Andaloussi SE, Schiffelers RM, Wood MJA, Vader P (2020) A CRISPR-Cas9-based reporter system for single-cell detection of extracellular vesicle-mediated functional transfer of RNA. Nat Commun 11:1113
Endo M, Iwakami S, Toki S (2021) Precision genome editing in plants via gene targeting and subsequent break-induced single-strand annealing. Plant Biotechnol J 19:563–574
Erikson O, Hertzberg M, Nasholm T (2004) A conditional marker gene allowing both positive and negative selection in plants. Nat Biotechnol 22:455–458
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu JK (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111:4632–4637
Fiorenza MT, Bevilacqua A, Bevilacqua S, Mangia F (2001) Growing dictyate oocytes, but not early preimplantation embryos, of the mouse display high levels of DNA homologous recombination by single-strand annealing and lack DNA nonhomologous end joining. Dev Biol 233:214–224
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826
Gao X, Chen J, Dai X, Zhang D, Zhao Y (2016) An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol 171:1794–1800
Gisby MF, Mudd EA, Day A (2012) Growth of transplastomic cells expressing D-amino acid oxidase in chloroplasts is tolerant to d-alanine and inhibited by d-valine. Plant Physiol 160:2219–2226
Gürel F, Zhang Y, Sretenovic S, Qi Y (2020) CRISPR-Cas nucleases and base editors for plant genome editing. aBIOTECH 1:74–87
He Y, Zhu M, Wang L, Wu J, Wang Q, Wang R, Zhao Y (2018) Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol Plant 11:1210–1213
He Y, Zhao Y (2019) Technological breakthroughs in generating transgene-free and genetically stable CRISPR-edited plants. aBIOTECH 1:88–96
Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M (2004) ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol 55:389–398
Kong X, Pan W, Sun N, Zhang T, Liu L, Zhang H (2021) GLABRA2-based selection efficiently enriches Cas9-generated nonchimeric mutants in the T1 generation. Plant Physiol 187:758–768
Lao K, **ao Y, Huang Q, Mo B, Dong X, Wang X (2023) Establishment of an efficient early flowering-assisted CRISPR/Cas9 gene-editing system in Arabidopsis. Plant Cell Rep 42:211–214
Lim S, Woo HJ, Lee SM, ** YM, Cho H (2007) d-amino acid oxidase (DAO) gene as a novel selection marker for plant transformation. J Plant Biotechnol 34:31–36
Liu Y, Zeng J, Yuan C, Guo Y, Yu H, Li Y, Huang C (2019) Cas9-PF, an early flowering and visual selection marker system, enhances the frequency of editing event occurrence and expedites the isolation of genome-edited and transgene-free plants. Plant Biotechnol J 17:1191–1193
Lu HP, Liu SM, Xu SL, Chen WY, Zhou X, Tan YY, Huang JZ, Shu QY (2017) CRISPR-S: an active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol J 15:1371–1373
Oh JM, Myung K (2022) Crosstalk between different DNA repair pathways for DNA double strand break repairs. Mutat Res Genet Toxicol Environ Mutagen 873:503438
Pilone MS (2000) d-amino acid oxidase: new findings. Cell Mol Life Sci 57:1732–1747
Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G (2007) Physiological functions of d-amino acid oxidases: from yeast to humans. Cell Mol Life Sci 64:1373–1394
Pollegioni L, Sacchi S, Murtas G (2018) Human d-amino acid oxidase: structure, function, and regulation. Front Mol Biosci 5:107
Rinne J, Witte CP, Herde M (2021) Loss of MAR1 function is a marker for co-selection of CRISPR-induced mutations in plants. Front Genome Ed 3:723384
Stinson BM, Loparo JJ (2021) Repair of DNA double-strand breaks by the nonhomologous end joining pathway. Annu Rev Biochem 90:137–164
Tang X, Ren Q, Yang L, Bao Y, Zhong Z, He Y, Liu S, Qi C, Liu B, Wang Y, Sretenovic S, Zhang Y, Zheng X, Zhang T, Qi Y, Zhang Y (2019) Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol J 17:1431–1445
Tian Y, Zhong D, Li X, Shen R, Han H, Dai Y, Yao Q, Zhang X, Deng Q, Cao X, Zhu JK, Lu Y (2023) High-throughput genome editing in rice with a virus-based surrogate system. J Integr Plant Biol 65:646–655
Tsutsui H, Higashiyama T (2017) pKAMA-ITACHI vctors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol 58:46–56
Vu TV, Das S, Nguyen CC, Kim J, Kim JY (2022) Single-strand annealing: molecular mechanisms and potential applications in CRISPR-Cas-based precision genome editing. Biotechnol J 17:e2100413
Wang J, Chen H (2020) A novel CRISPR/Cas9 system for efficiently generating Cas9-free multiplex mutants in Arabidopsis. aBIOTECH 1:6–14
Wang ZP, **ng HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16:144
Wolabu TW, Park JJ, Chen M, Cong L, Ge Y, Jiang Q, Debnath S, Li G, Wen J, Wang Z (2020) Improving the genome editing efficiency of CRISPR/Cas9 in Arabidopsis and Medicago truncatula. Planta 252:15
**e K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112:3570–3575
**e X, Ma X, Zhu Q, Zeng D, Li G, Liu YG (2017) CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Mol Plant 10:1246–1249
Xu W, Yang Y, Liu Y, Kang G, Wang F, Li L, Lv X, Zhao S, Yuan S, Song J, Wu Y, Feng F, He X, Zhang C, Song W, Zhao J, Yang J (2020a) Discriminated sgRNAs-based surrogate system greatly enhances the screening efficiency of plant base-edited cells. Mol Plant 13:169–180
Xu R, Li J, Liu X, Shan T, Qin R, Wei P (2020b) Development of plant prime-editing systems for precise genome editing. Plant Commun 1:100043
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Zhang Q, **ng HL, Wang ZP, Zhang HY, Yang F, Wang XC, Chen QJ (2018) Potential high-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention. Plant Mol Biol 96:445–456
Acknowledgements
The authors thank Qi-Jun Chen for the pHEE401E construct. This work was supported by the National Key Research and Development Program of China (grant 2019YFA0906202) to J.-F.L., the National Natural Science Foundation of China (grants 31900305 and 32370294) and the Natural Science Foundation of Guangdong Province (grant 2020A1515010465) to F.-Z.W.
Author information
Authors and Affiliations
Contributions
JFL conceived the study. FZW and YB conducted the experiments. FZW, XX, and ZL analyzed the data. JFL and FZW wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, FZ., Bao, Y., Li, Z. et al. A dual-function selection system enables positive selection of multigene CRISPR mutants and negative selection of Cas9-free progeny in Arabidopsis. aBIOTECH 5, 140–150 (2024). https://doi.org/10.1007/s42994-023-00132-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42994-023-00132-6