Abstract
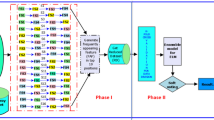
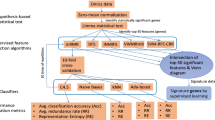
Over the past few decades, there has been a massive growth in the volume of biological data. In such datasets, the influence of dimensionality bias or existence of repetitive, noisy, and irrelevant genes has become a severe barrier in classifying gene expression data. Therefore, to reduce the impact of noisy genes and precisely identify gene patterns for enhancing classification accuracy, feature selection strategies are employed. This article proposes an innovative hybrid feature selection model by mixing statistical and filter-feature selection methodologies. Following the initial step of normalizing each sample, a non-parametric Kruskal–Wallis (KW’s) test and Bonferroni Correction (BC) using together to pick relevant genes. Finally, a correlation-based feature selection (CFS) method employed to determine how different genes are related, and a greedy search policy used to eliminate repetitious genes to discover promising gene subsets. Based on the results and comparison of six distinct microarray datasets, it is clear that the proposed method is superior to Chi-square, Joint Mutual Information (JMI), Conditional Mutual Information Maximization (CMIM), Relief-F, and Minimum Redundancy Maximum Relevance (mRMR) state-of-the-art feature selection algorithms while using Support Vector Machine (SVM), Naïve Bayes (NB), K-Nearest Neighbors (k-NN), and Decision Tree (DT) classifiers respectively. These findings lead us to believe that the suggested feature selection algorithm can effectively discriminate cancer patients from healthy persons.







Similar content being viewed by others

References
Jones PA, Baylin SB (2007) The epigenomics of cancer. Cell 128(4):683–692
Brown PO, Botstein D (1999) Exploring the new world of the genome with dna microarrays. Nature genetics 21(1):33–37
Lockhart DJ, Winzeler EA (2000) Genomics, gene expression and dna arrays. Nature 405(6788):827–836
Tinker AV, Boussioutas A, Bowtell DD (2006) The challenges of gene expression microarrays for the study of human cancer. Cancer cell 9(5):333–339
Ang JC, Mirzal A, Haron H et al (2015) Supervised, unsupervised, and semi-supervised feature selection: a review on gene selection. IEEE/ACM Trans Comput Biol Bioinform 13(5):971–989
Saha S, Biswas S, Acharyya S (2016) Gene selection by sample classification using k nearest neighbor and meta-heuristic algorithms. In: 2016 IEEE 6th international conference on advanced computing (IACC), IEEE, pp 250–255
Deng L, Pei J, Ma J et al (2004) A rank sum test method for informative gene discovery. In: Proceedings of the tenth ACM SIGKDD international conference on Knowledge discovery and data mining, pp 410–419
Liao C, Li S, Luo Z (2006) Gene selection using wilcoxon rank sum test and support vector machine for cancer classification. In: international conference on computational and information science, Springer, pp 57–66
Ma J, Li F, Liu J (2005) Non-parametric statistical tests for informative gene selection. In: International Symposium on Neural Networks, Springer, pp 697–702
Das U, Hasan MAM, Rahman J (2019) Influential gene identification for cancer classification. In: 2019 International Conference on Electrical. Computer and Communication Engineering (ECCE), IEEE, pp 1–6
Chandra B, Gupta M (2011) An efficient statistical feature selection approach for classification of gene expression data. J Biomed Inform 44(4):529–535
Lu X, Peng X, Liu P et al (2012) A novel feature selection method based on cfs in cancer recognition. In: 2012 IEEE 6th International Conference on Systems Biology (ISB), IEEE, pp 226–231
Sharma M (2019) Improved autistic spectrum disorder estimation using cfs subset with greedy stepwise feature selection technique. Int J Inform Technol:1–11
Vanitha CDA, Devaraj D, Venkatesulu M (2015) Gene expression data classification using support vector machine and mutual information-based gene selection. Procedia Comput Sci 47:13–21
Juneja K, Rana C (2020) An improved weighted decision tree approach for breast cancer prediction. Int J Inform Technol 12(3):797–804
Rajab M, Wang D (2020) Practical challenges and recommendations of filter methods for feature selection. J Inform Knowl Manag 19(1):2040019
Zhang Y, Ding C, Li T (2008) Gene selection algorithm by combining relieff and mrmr. BMC Genom 9(2):1–10
Wang A, An N, Chen G et al (2015) Accelerating wrapper-based feature selection with k-nearest-neighbor. Knowl Based Syst 83:81–91
Tabakhi S, Najafi A, Ranjbar R et al (2015) Gene selection for microarray data classification using a novel ant colony optimization. Neurocomputing 168:1024–1036
Morovvat M, Osareh A (2016) An ensemble of filters and wrappers for microarray data classification. Mach Learn Appl An Int J 3(2):1–17
Sasikala S, Alias Balamurugan SA, Geetha S (2016) Multi filtration feature selection (mffs) to improve discriminatory ability in clinical data set. Appl Computi Inform 12(2):117–127
Wang A, An N, Yang J et al (2017) Wrapper-based gene selection with markov blanket. Comput Biol Med 81:11–23
Su Q, Wang Y, Jiang X et al (2017) A cancer gene selection algorithm based on the ks test and cfs. BioMed research international 2017
Rouhi A, Nezamabadi-pour H (2017) A hybrid feature selection approach based on ensemble method for high-dimensional data. In: 2017 2nd Conference on Swarm Intelligence and Evolutionary Computation (CSIEC), IEEE, pp 16–20
Ke W, Wu C, Wu Y et al (2018) A new filter feature selection based on criteria fusion for gene microarray data. IEEE Access 6:61065–61076
Jansi Rani M, Devaraj D (2019) Two-stage hybrid gene selection using mutual information and genetic algorithm for cancer data classification. J Med Syst 43(8):1–11
Shukla AK, Tripathi D (2020) Detecting biomarkers from microarray data using distributed correlation based gene selection. Genes Genom 42(4):449–465
Shukla AK, Pippal SK, Gupta S et al (2020) Knowledge discovery in medical and biological datasets by integration of relief-f and correlation feature selection techniques. J Intell Fuzzy Syst 38(5):6637–6648
Dass S, Mistry S, Sarkar P et al (2021) An optimize gene selection approach for cancer classification using hybrid feature selection methods. In: International Conference on Advanced Network Technologies and Intelligent Computing, Springer, pp 751–764
Halim Z et al (2021) An ensemble filter-based heuristic approach for cancerous gene expression classification. Knowl Based Syst 234(107):560
Sharma A, Mishra PK (2022) Performance analysis of machine learning based optimized feature selection approaches for breast cancer diagnosis. Int J Inform Technol 14(4):1949–1960
Han J, Pei J, Tong H (2022) Data mining: concepts and techniques. Morgan kaufmann
Sarwar A, Ali M, Manhas J et al (2020) Diagnosis of diabetes type-ii using hybrid machine learning based ensemble model. Int J Inform Technol 12(2):419–428
Cano A, Masegosa A, Moral S (2005) ELVIRA biomedical data set repository (Online). http://leo.ugr.es/elvira/DBCRepository/
Acknowledgements
We owe a debt of gratitude to the Computer Science and Engineering department of MAKAUT, West Bengal for their unwavering support. We’d also like to thank the department of In Vitro Carcinogenesis and Cellular Chemotherapy of CNCI, Kolkata for their support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dass, S., Mistry, S., Sarkar, P. et al. A proficient two stage model for identification of promising gene subset and accurate cancer classification. Int. j. inf. tecnol. 15, 1555–1568 (2023). https://doi.org/10.1007/s41870-023-01181-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41870-023-01181-2