Abstract
Hospital databases provide complex data on individual patients, which can be analysed to discover patterns and relationships. This can provide insight into medicine that cannot be gained through focused studies using traditional statistical methods. A multivariate analysis of real-world medical data faces multiple difficulties, though. In this work, we present a methodology for medical data analysis. This methodology includes data preprocessing, feature analysis, patient similarity network construction and community detection. In the theoretical sections, we summarise publications and concepts related to the problem of medical data, our methodology, and rheumatoid arthritis (RA), including the concepts of disease activity and activity measures. The methodology is demonstrated on a dataset of RA patients in the experimental section. We describe the analysis process, hindrances encountered, and final results. Lastly, the potential of this methodology for future medicine is discussed.
Similar content being viewed by others
Introduction
Modern medicine utilises many analysis methods that generate large quantities of data. Most of the time, this data is used in narrow, focused views of a single medical doctor in a context of a single disease. However, the data is stored electronically in hospital databases and can thus be conglomerated into complex views of individual patients. These massive datasets of seemingly incoherent data can provide new insight into medicine, not attainable by the traditional focused approach. To connect the knowledge of isolated biological processes, medical research began to view them as complex networks (Alm and Arkin 2003). Despite advancements in medical data storage, data collection has severe shortcomings. Work overload, typographical and copy-paste errors, and mistaken autocorrections are some of the culprits of the poor state of many datasets (Turchin et al. 2011; Lewis 2021). Since these problems prevail (Abeysooriya et al. 2021), data analysis methodology must adapt to overcome them.
This work presents a methodology for analysing medical data based on similarity networks. The workflow consists of data preprocessing and feature analysis, preparing partial datasets of selected features and an iterative feature reduction until a quality network with information relevant to the domain is found. The similarity network is then constructed and analysed. In “Experiment” section, the method is demonstrated using the LRNet algorithm based on local representativeness analysis (Ochodkova et al. 2017) on a dataset of rheumatoid arthritis (RA) patients.
Disease activity in rheumatoid arthritis
RA is a heterogeneous autoimmune inflammatory disease characterised by joint swelling and tenderness. The cornerstone of RA patients’ management and treatment is disease activity. Contemporary RA treatment follows a treat-to-target paradigm (Smolen et al. 2016), where the patient is treated proactively and dynamically while disease activity changes are closely monitored. This allows for an effective treatment with a minimal biological burden. Disease activity is quantified using activity measures, with the most prominent ones being DAS28 (van Gestel et al. 1998), SDAI (Smolen et al. 2003), and CDAI (Aletaha et al. 2005). DAS28 (Disease Activity Score - 28 joints counts) is considered the golden standard (van der Heijde and Jacobs 1998; Boers et al. 1994) by both the American College of Rheumatology (ACR) and the European Alliance of Associations for Rheumatology (EULAR). DAS28 lies in a sweet spot between precision and clinical feasibility (Landewé et al. 2006). Thus, it was used as the ground truth in establishing the other activity measures, in comparing them (Leeb et al. 2005) and in this article.
The activity measures are calculated using biological markers and symptom quantifications but vary in the specific parameters used as well as the calculation formulae. The components usually include peripheral blood inflammatory markers (C-reactive protein, CRP; erythrocyte sedimentation rate, ESR), affected joint counts (swollen joints count, SJC; tender joints count, TJC) and questionnaires inspecting the patient’s general health (GH). A typical pattern in the activity measure formulas is that multiple factors add to the activity score, as seen in the DAS28-CRP formula:
The resulting activity score is then assigned to one of four activity groups (I–IV, low to high activity). These represent the groups of patients with distinct treatment strategies. Group I contains patients in remission or with low disease activity. Patients in group II are under surveillance. Patients in group III begin active treatment, and patients in group IV are treated intensively. Since patients in individual groups require different amounts of attention from the clinician, this categorization can be simplified to non-active patients (groups I+II) and active patients (groups III+IV).
The additive nature of the activity measures causes heterogeneity in the activity groups, where patients with different parameter values can be assigned the same activity score value. Additional sources of inaccuracy are the general health questionnaires because of their subjective nature. Both the general health questionnaire filled by the patient and another one filled by the rheumatologist are subject to bias (Zhang et al. 2021; Pincus and Morley 2001).
This article is organised as follows: in “Related work and methods” section, we discuss related work; in “Methodology” section, we propose a methodology for medical data analysis and visualisation; in “Experiment” section, we demonstrate the methodology on a RA patients dataset; “Conclusions” section concludes the article.
Related work and methods
Network science in RA
Clustering analysis is less prevalent in RA studies compared to traditional statistical methods. Still, multiple studies utilizing clustering and network analysis methods were published recently. One study (Lee et al. 2014) used hierarchical agglomerative clustering to study pain in RA patients. A more recent study (Jung et al. 2021) used a combination of k-means clustering and tSNE algorithm (Van der Maaten and Hinton 2008) to study the risk of biological treatment initiation in RA patients. Another study (Platzer et al. 2022) used k-means clustering for longitudinal data to study radiological damage trajectory curves. Yet another study (Crowson et al. 2022) analyzed RA comorbidity factors to compare the clustering performance of hierarchical and k-means clustering with network analysis (with Cramer’s V distance as a dissimilarity measure).
Network analysis has been used in several studies concerning RA, mostly analysing gene co-expression (Zheng and Rao 2015; Ma et al. 2017; **ao et al. 2021; Yadalam et al. 2022). Network analysis was also used to study RA drug-target interactions (Zhang et al. 2015; Song et al. 2019) and in RA metastudies studying common comparators of disease activity or treatment (Zhu et al. 2023; Budtarad et al. 2023). All these studies used data readily in a network form. As per our knowledge, our study is the first to use a network analysis approach to vector data in RA.
Network construction
Network science meets various applications, from social media and marketing to gene co-expression and protein modelling. In some of these applications, the data form a network, e.g. social networks (Chakraborty et al. 2018) or biological networks (Pavlopoulos et al. 2011) In these cases, network-based analysis methods can be directly applied. However, in other applications, such as coexpression modelling (Xu et al. 2002) or spectral clustering (Jia et al. 2014), the data needs to be transformed from vector data to network data before applying network analysis methods. Network construction is often necessary when solving problems arising from machine learning applications since vector data captures less information than network data. The most important information captured by networks is structural information about data relationships.
Constructing a network from vector data is a two-part process. First, the similarity (or dissimilarity) between all pairs of records is calculated using a method designed to produce a similarity (or dissimilarity) matrix. Some of the measures used are described below in the following subsection. The second part of the network construction process is sparsification, which aims to select the edges important to the constructed network.
Patient similarity networks
A patient similarity network (PSN)(Wang et al. 2014) is a network structure that comprises vertices (nodes) representing patients and weighted undirected edges representing the patients’ similarity. PSNs have been previously used to analyse patient data and to support the visualization of trends (Turcsanyi et al. 2019; Manukyan et al. 2020; Gallo et al. 2020; Mikulkova et al. 2021). One advantage of PSNs is the ease of use in multivariate analysis. The agility of the method comes from the ability to easily change the feature set used to construct the PSN and to visualize the distribution of features within the network without influencing the structure of the PSN. Additionally, PSNs have great utility in visualization, providing easily readable and understandable information to domain experts without requiring deep knowledge of network science. An example of a patient similarity network is depicted in Fig. 1.
(Dis)similarity measures
In standard notation, a network G(V, E, W) comprises a set of V vertices (nodes) representing data records, a set of (directed or undirected) edges E connecting the vertices, and a set of edge weights W. This weight usually represents common events (e.g. publications, event participation, gene co-expression), or a calculated distance or similarity between the vertices. There are many distance and similarity measures, and they all come with their advantages and caveats.
Euclidean distance is suitable for numerical data, in which the actual distance of records carries practical information. However, euclidean distance is unsuitable for high-dimensional data, where Manhattan distance is more appropriate (Aggarwal et al. 2001). Mahalanobis distance (Mahalanobis 1936) is used when the axes of n-dimensional space are not independent (redundancy in data features). Cosine similarity is used for text data and when the similarity of record proportions is of interest. There are other similarity measures used in specific applications, e.g. Jensen-Shannon distance (Endres and Schindelin 2003) for distributions, Hamming distance (Hamming 1980) for binary messages, Jaccard index (Jaccard 1901) for sets, and neighbourhood similarity measure for videos (Wang et al. 2009). Gaussian kernel (Belkin et al. 2006) is a non-linear function of Euclidean distance. This attribute is practical in network analysis and spectral clustering since Gaussian similarity can directly translate to edge weight (Nataliani and Yang 2019; Ng et al. 2001).
The selection of an ideal distance or similarity measure is thus a problem-specific issue. Moreover, network construction is further complicated by specific attributes the vector data associated with the nodes may have: data lie in a high-dimensionality space; data are derived from different distributions; data is contaminated by noise; data are probabilistic (Qiao et al. 2018).
Sparsification
However, creating links between pairs of vertices with weights according to the received (dis)similarity matrix by simply preserving edges with all calculated (dis)similarities leads to (almost) complete networks, which is not desirable. We need a high-quality network that ideally contains a giant component and is as sparse as possible to capture the relationships between vertices. Thus, sparsification is important because it improves accuracy and robustness to noise (Silva et al. 2018). We consider noise as links with small values (or large values in the case of dissimilarity) that can distort the resulting network topology. Thus, noise removal is the second important step in constructing networks from vector data.
Traditional sparsification methods include the following (Belkin and Niyogi 2003):
-
Generally, a k-nearest neighbour (k-NN) network is directed. An edge from \(v_i\) to \(v_j\) exists if and only if \(v_j\) is among the k most similar elements to \(v_i\) (or among the k least distant elements in the case of dissimilarity, i.e. distance). This approach requires ordering the rows of the matrix. After sorting, for row i, links are established between vertex i and the first k items in the sorted list corresponding to the elements in the i-th row of the matrix.
-
\(\epsilon\)-radius threshold method - an undirected edge between two nodes is added if the distance between them is smaller than \(\epsilon\), where \(\epsilon > 0\) is a predefined constant.
-
A combination of k-nearest neighbors and \(\epsilon\)-radius (Silva and Zhao 2012) - both methods are combined based on the density of particular network areas. In sparse areas, we use k-nearest neighbours; in dense areas, we use the \(\epsilon\)-radius technique.
Our work uses the Gaussian kernel similarity measure and the LRNet algorithm to transform the vector data into a network. As Zehnalova et al. showed in previous works (Zehnalova et al. 2014a, b), data reduction based on nearest neighbour and x-representativeness of nodes provides better results than sampling in preserving structures within a dataset. Authors of Ochodkova et al. (2017) proposed algorithm LRNet based on local representativeness, displaying a similar data structure preservation capability as was seen in the work by Zehnalova et al. The algorithm evaluates the nearest neighbours and the importance of individual nodes within the network structure while not requiring the manual setting of parameters.
Community detection
Community detection algorithms are used to reveal structures within a similarity network. The terms “cluster” and “community” are similar in their meaning, both denoting a group of closely located or connected records (i.e. patients in the context of medical data). Both terms are used interchangeably in this work, depending on the context. The community detection problem is similar to the more general clustering problem. In clustering, multiple attributes (features) are considered, while in community detection, the nodes’ features are already transformed into (weighted) edges. Traditional clustering methods such as hierarchical clustering (Scott 2000) are not well suited for network data. They tend to successfully find parts of communities that correspond to highly similar records but leave some records unassigned (Newman 2004). Methods devised for network community detection are based on different approaches: modularity-based methods (Newman and Girvan 2004; Blondel et al. 2008), Bayesian methods (Yan 2016), statistical-test-based methods (Wang and Bickel 2017), and spectral clustering methods (Saade et al. 2014), to name some. Another important aspect to consider is the difference between disjoint and overlap** communities, as the solutions to these problems differ fundamentally. More information on the problem of overlap** communities, where a node can be a member of multiple communities (typically social networks), can be found in Vieira et al. (2020). Medical data does not form overlap** communities since the patients are usually divided into disjoint groups.
Our methodology utilizes the Louvain algorithm (Blondel et al. 2008). This method is based on the optimization of modularity, that is, the relative density of the edges inside communities to the edges outside communities. It works in two-step iterations, where it first assigns nodes to communities and then tries to move singular nodes to different communities and observes how the modularity of the whole network changes. The solution with the highest positive modularity change is selected as a starting point for the next iteration. Louvain algorithm is typically used for large networks but has shown good results in small-scale medical data networks (Matta et al. 2023; Trajerova et al. 2022; Turcsanyi et al. 2019). The advantage of the Louvain algorithm is that it is fully automated and requires no settings. A disadvantage is that the order of node moving can influence the outcome, and small fluctuations in solutions can be observed. The authors report these differences do not significantly influence the resulting modularity.
Clustering evaluation
Clustering evaluation is elemental in assessing clustering or community detection results. There are two distinct approaches to cluster evaluation: external and internal measures (Theodoridis and Koutroumbas 2009).
External evaluation measures, e.g. Adjusted Rand Index (Hubert and Arabie 1985), Normalized Mutual Information (Chen et al. 2010), and Jaccard index (Jaccard 1912), compare the clustering solutions with a pre-specified structure, i.e. a ground truth (external information not included among the clustered features), which reflects the known facts about the dataset, domain knowledge or expert intuition (Halkidi et al. 2002).
Internal evaluation measures are based on data vectors within the dataset. One group of internal measures, such as Dunn index (Dunn 1974), Davies-Bouldin index (Davies and Bouldin 1979), or Silhouette score (Rousseeuw 1987) that assesses the similarity (fit) between the clusters. Internal measures choose a representative point within detected clusters and use compactness and separation measures to evaluate the solution based on the distances between these points (Pascual et al. 2010). Another approach used, e.g. by the gap statistic (Tibshirani et al. 2001), assesses the explained variation of solutions (so-called “knee-of-the-curve method”). Another group of internal measures (Ben-Hur and Guyon 2003; Bel Mufti et al. 2005) assesses the stability of the behaviour of the solution. These measures define a variability measure of clustering solutions and then perform clustering on samples taken from the dataset, searching for a solution showing the lowest variation of variability measure in the sampled datasets.
A different approach to cluster evaluation is found in graph-based evaluation measures. In these, the data is first transformed into a network, which is then assessed by graph-based measures like modularity (Newman and Girvan 2004) evaluating the quality of a division of the graph, and conductance (Kannan et al. 2004) weighing groups of nodes by their importance.
Multiple studies reported no single best evaluation measure (Dimitriadou et al. 2002; Maulik and Bandyopadhyay 2002; Arbelaitz et al. 2013). A recent work (Spalenza et al. 2021) presented an algorithm combining internal and external evaluation measures, achieving significant improvements in evaluation. A similar approach is presented in our work.
Methodology
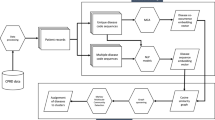
In this section, we propose a methodology for real-world medical data analysis. The workflow is depicted in Fig. 2.
Data preprocessing
As stated before, real-world medical data have many problems that need to be addressed before the analysis: typos detection, unification of varying values representing one actual state, removal of duplicated features, and remediation of incorrect Excel autocorrections. This step requires considerable insight into the domain, and thus it is advisable to involve a domain expert. Then the features are analysed using descriptive statistical methods and preprocessed according to detected distributions and their abnormalities. Features with lognormal distribution are transformed with a logarithm. These transformations, in particular, should be discussed with the domain expert since many features might have characteristics originating in the data collection method. A correct transformation is necessary to preserve these characteristics, e.g. DNA amplification primary data need to be transformed specifically with \(log_2\). Then, features are normalized by rescaling to the interval [0, 1]. Finally, the presence of potential outliers is resolved.
(Dis)similarity measure selection
The measure selection is based both on the data structure, discussed in “Related work and methods” section, and on what the individual features represent and their domain relevance.
Preliminary network analysis
With a preprocessed dataset and a similarity measure selected, a preliminary network is constructed with data vectors composed of all features. This network is typically very dense, and the data structures are not discernible. However, the importance and influence of individual features can be judged based on this network. The Pearson correlation coefficient between individual feature pairs is used to assess the redundancy of features. A domain expert is vital for this step, as the significant value of a correlation coefficient is domain-specific.
Preparing feature sets
After removing features of high redundancy or low significance, features are grouped into feature sets. These are subsets of data comprising at most ten features, selected based on domain-knowledge-based preference and feature significance. The feature sets are then inspected individually.
Feature sets analysis by patient similarity networks
Individual feature sets are analysed using patient similarity networks. Iterations of the following steps are performed on each feature set:
-
1.
Network construction
-
2.
Network quality assessment
-
3.
Network visualization and feature analysis
-
4.
Feature removal
Network construction
A network is constructed using the LRNet algorithm (Ochodkova et al. 2017). However, a different network construction algorithm may be used instead without impacting the rest of the methodology. The quality of the network is assessed based on four criteria: the number of detected communities, the modularity of the network, the embeddedness of the communities, and the silhouette of the communities. Additionally, the distance of community centroids is considered. The centroids effectively represent an average patient of said community. A quality network would ideally yield distant centroids and thus cluster patients into groups with distinct feature profiles.
Network quality assessment
A network with a high number of small clusters is discarded, as too few clusters indicate insufficient intra-feature diversity. However, this happens scarcely, thanks to the feature pre-selection step. Network modularity represents the mutual separation of clusters. Embeddedness (Hric et al. 2014) represents the interconnectedness of nodes within clusters. The silhouette score of a node represents the ambiguity of its assignment to its cluster, with a low silhouette score indicating that the node is similar to not one but multiple clusters. Thus, the ideal network is highly modular, with an optimal number of highly embedded clusters with distant centroids and nodes with high silhouette scores.
A high-quality network can be of little value should the information provided be irrelevant to the aim of the analysis (clinical significance). This can be assessed by the domain expert or by inspecting the detected clusters and a class feature, provided one is present in the dataset. This class feature must not be part of the network construction feature set. The assessment is always binary, comparing one class within a cluster against the rest of the data. That is, objects of the class within the cluster are true positives, and any other objects therein are false positives. Analogously, class objects outside the cluster are false negatives, and any other objects therein are true negatives.
A dominant class is determined by assessing all classes and comparing their precision and recall values for individual cluster-class pairs. \(F_1\) score, which balances the importance of precision and recall by a harmonic mean, can provide more complex information. Since the classes and clusters are usually imbalanced, multi-class Mathew’s correlation coefficient can be used to provide more understanding.
A good cluster then contains a single decisively dominant class (high precision) and a decisive majority of this class’s objects (high recall).
It should be noted, though, that this methodology aims not to find the optimal solution but rather to find a good enough solution that would be easily understandable and highly informative to the clinician. Thus, there can be multiple acceptable outcomes of this analysis.
Network visualization and feature analysis
Not every feature set has the potential to reveal data structures meaningful in the context of the domain. After obtaining a quality network, a discussion with a domain expert is necessary to assess its clinical value. For this discussion, an easily understandable visualization is necessary. This can be produced using any preferred network visualization software. The experiment in “Experiment” section utilizes Gephi software (Bastian et al. 2009) with layout algorithm ForceAtlas 2 (Jacomy et al. 2014).
Removing features
Inspecting the distribution of individual features in relation to the detected communities together with the domain expert leads to adding, removing and/or replacing individual features from promising feature sets. This may lead to changes in the network properties, especially in smaller feature sets.
Final visualization
When a satisfactory network is found, a more profound visualization step follows. First, the network layout is revised so that nodes do not overlap, and any structures too dense to discern are adjusted. This can be done automatically by modifying the parameters of the used network layout algorithm (e.g. gravity, repulsion, network sparsification, overlap prevention) or by manually adjusting the position of the nodes. Since the similarity information is carried by the edges and not by the nodes’ location, minor layout adjustments that improve the overall readability of the network are not detrimental.
Second, a visualization of “structural features” (the values which form the data vectors used in network construction) is prepared by portraying the distribution of individual features within the network. This allows for an easy understanding of how groups of patients (detected clusters) fundamentally differ. Third, a visualization of additional features is prepared. These features do not participate in the network construction, but showing their distribution within the network reveals the relationship between them and the patient groups.
Using these visualizations, the domain expert then assesses the results and determines their clinical significance.
Experiment
The Department of Internal Medicine III - Nephrology, Rheumatology and Endocrinology, University Hospital Olomouc and the Department of Immunology, Palacky University Olomouc, provided the dataset of RA patients for the experimental part. All patients within the dataset provided informed consent to use their data. Ethical committees of both the University Hospital and Palacky University approved the study. The RA dataset comprised 78 patients and 205 features, spread among three data files (clinical, protein, and blood count data) with inconsistent formats. This and other issues were solved during the data preprocessing.
The experiment aimed to find a set of RA patient features that would reach similar results of patient classification by disease activity as DAS28. There were three requirements for the task:
-
1.
to use exclusively objective data (i.e. not to use general health questionnaires)
-
2.
to use patient features routinely collected during the regular checkups (so that there are no or minimal changes in the clinical practice)
-
3.
to include blood lipid parameters into the selected features (based on the domain knowledge of the clinicians)
Data preprocessing
First, the data files were joined, and the format was unified. The data types and unique values were inspected to remove redundant values and unify the format of individual features. These redundancies were introduced by the collaboration of multiple healthcare staff on the clinical data collection and by an overlap of clinical and blood count data. Especially problematic was the influence of Microsoft Excel’s autocorrect function on several features, where multiple numerical or textual values were changed into date values. Direct access to medical records (available due to a close collaboration with the clinicians) was invaluable in resolving these issues and data conflicts.
Based on a discussion with a clinician and the task requirements, feature pre-selection was performed. Ultimately, two patients and 52 features were removed because of missing data or irrelevance to the task. In the preprocessed dataset, the normality of data was assessed, and where necessary, the features were normalized: e.g. logarithmic transformation was applied to protein features (\(log_2\)) and CRP (\(ln(1+x)\)).
A clinician established a ground-truth feature by grou** disease activity groups into non-active patients (groups I+II) and active patients (groups III+IV).
(Dis)similarity measure selection
After inspection of the final dataset and discussion with the clinician, the Gaussian kernel was selected as a similarity measure:
where \(\sigma ^2\) is variance and \(\sigma\) was set to 0.5.
Feature sets analysis
LRNet algorithm (Ochodkova et al. 2017) using Gaussian kernel similarity measure was used for patient similarity network construction. A preliminary network was constructed to assess the significance of features. Multiple feature sets were prepared based on an analysis of this network and the feature selection factors (see “Methodology” section). Each of these sets focused on different key features recommended by the clinicians. Special attention was paid to blood lipid level parameters, as requested.
First, sets focusing on the components of activity measures were analyzed. The well-known correlation between ESR and CRP (Kotulska et al. 2015) was confirmed with Pearson correlation coefficient and matching trends observed in produced networks (Fig. 3). A correlation between the SJC and TJC was found (Fig. 3), corresponding with the reported correlation of both features with the disease activity (Riazzoli et al. 2010). Because of the high redundancy in these two pairs of features, only CRP and SJC were selected for further analyses based on their higher significance in the similarity network.
Second, sets focusing on the protein data features were analyzed. This revealed a significance of blood level of proteins CXCL10, CXCL9, CCL20, CCL7, IL-8, sOPG, and CXCL1. Another group of proteins (VEGFA, sLIFR, sIL-15RA, sCD40, and CSF1) also provided a quality network. Both networks were later discarded as clinically insignificant due to a low resolution of disease activity classification. Third, sets focusing on the clinical data were analyzed, but these did not provide relevant results. Last, the feature sets containing blood lipid features with variations of other features were inspected. The results are presented in the following subsection.
Final visualisation and results
The final result was reached in a network constructed using SJC and CRP, total cholesterol, LDL-cholesterol, and triglycerides blood levels. This result confirmed the importance of blood lipid parameters, as their addition to the network features provided considerably better results in disease activity classification than any previous feature combinations. The experiment also shows that disease activity cannot be reduced to a single parameter and requires a multivariate analysis approach.
Four patient communities were detected in the network (Fig. 4). The communities were compared to the distribution of DAS28 disease activity groups (Fig. 5). The green community contained exclusively active patients (activity groups III+IV). The red community contained predominantly active patients (88%). The blue community contained roughly the same amount of active (III+IV) and non-active (I+II) patients (47% active). The orange community contained predominantly non-active patients (11% active).
The clustering quality for the established ground-truth feature (non-active vs active patients) was assessed using precision, recall, and \(F_1\) score. The values for dominant classes in individual clusters as depicted in Table 1. Figures 6, 7, 8, 9 and 10 show the distribution of the features within the network.
As is shown in Fig. 11, the profiles of the community centroids were distinct, defining profiles of average patients of the respective communities.
Several patients with activity scores different from their neighbours were discovered. Consulting a clinician revealed that three of these patients suffered from a concurrent disease or were on a medication which altered their blood lipid profiles (Fig. 12). These facts were not included in the clinical data provided.
Experiment conclusions
From the clinical point of view, the experiment found a set of objective features in RA patients that can be utilized in assessing disease activity with a performance similar to that of disease activity measures. Unlike disease activity measures, these features include easily obtainable blood lipid measures and do not include general health questionnaires. The experiment shows that disease activity in its complexity cannot be reduced to a single parameter, and multivariate analysis proves to be a good approach.
Additionally, the experiment revealed two groups with distinct profiles within high-activity patients and a group of active patients with very low swollen joint counts, neither of which was described before. Furthermore, the experiment revealed multiple patients with feature profiles strikingly different from their neighbours. It was found later that the features of these patients were influenced by external factors (other concurrent diseases and treatments) not included within the dataset. This shows that patient similarity analysis can reveal information about the patients not obtainable by the standard approach of the domain.
From the data science point of view, the experiment verified the utility of the methodology and demonstrated the usefulness of visualization of patient data as patient similarity networks. This application also shows a potential for creating a software tool for clinical practice. In such a tool, a clinician would add a new patient to the network, which would be recalculated and reconstructed. In the updated network, the patient is placed near other similar patients. Since the clinician already has extensive knowledge of these neighbours, they can make better decisions regarding the new patient.
With a large enough dataset, this tool could find use in the emerging era of telemedicine. This term encompasses any medical activity involving an element of distance (Wootton 2001) and, in a broader sense, the “backroom” processes of automating the clinical process. This tool could include options for direct contact with patients that would allow for data collection from home and general practitioners without the need to visit a hospital. This increased feasibility would provide more quality, frequent, and complete data without burdening the medical staff and the patient.
Conclusions
In this work, we presented a methodology for medical data analysis based on patient similarity network analysis. We discussed the benefits of this approach and demonstrated the methodology on a dataset of RA patients. The experiment found a set of features that complied with the experiment requirements. The patient similarity network built using these features revealed four disjoint patient subsets with differing feature profiles. The experiment also demonstrated how the analysis of patient similarity could provide additional information beyond the experiment’s aim.
Furthermore, we demonstrated the utility of the presented methodology for cooperation between a data scientist and a domain expert (clinician), which we believe will be highly important in future medicine. With the trend of telemedicine, this application could lay a basis for a software tool that would help the clinical practice and allow for more feasible and thorough data collection. This, in turn, would be valuable for further progress and development.
Example of a patient similarity network. The nodes depict RA patients, and the edges depict their similarity. A Communities detected with the network. B Distribution of DAS28 score within the network on a four-point colour scale (dark blue, green, yellow, red). C Distribution of DAS28 activity groups I–IV within the network, coloured green, yellow, orange, and red, respectively
Distributions of correlating pairs of features. Correlating pairs are visualized in the final PSN in two columns: on the left, erythrocyte sedimentation rate and C-reactive protein; on the right, swollen and tender joint counts. A three-colour scale (blue-yellow-red) was found to provide good readability
Final PSN showing the distribution of DAS28 activity groups. Non-active patients are coloured green (group I) and yellow (group II). Active patients are coloured orange (group III) and red (group IV). The activity trend can be clearly visible rising from low values on the right side of the network towards high values on the left. The green community contains purely active patients (III+IV). The red community contains predominantly active patients (88%). The blue community is composed randomly of active (47%) and non-active (53%) patients. The orange community contains predominantly non-active patients (89%)
Distribution of CRP (C-reactive protein) in the final PSN. The distribution of CRP is visualized on a light-to-dark scale, with the high values concentrated on the left side of the network, highly correlated with the disease activity (compare to Fig. 5). Note that CRP was transformed as \(\ln {\left( CRP + 1\right) }\) during preprocessing
Distribution of SJC (swollen joints count) in the final PSN. The distribution of SJC is visualized on a light-to-dark scale. The distribution is correlated with the disease activity (compare to Fig. 5). A surprising profile of low SJC in patients with an elevated CRP is clearly visible within the blue community (compare to Fig. 6)
Distribution of LDL-cholesterol (low-density-lipoprotein cholesterol) in the final PSN. The distribution of LDL-cholesterol is visualized on a light-to-dark scale. The trend is visually very similar to that of total cholesterol (Fig. 8), but removing LDL-cholesterol from the network reduces its quality. This shows how a purely visual assessment fails to discern data in multivariate analysis
Centroid profiles of detected communities in the final PSN. While both red (active) and blue (mixed) communities display elevated CRP, the red community shows the lowest mean value in SJC of all communities despite many of its patients being considered active by DAS28. Four detected communities are coloured green, red, blue, and orange. CRP—C-reactive protein, SJC—swollen joints count, TC—total cholesterol, LDL-C - low-density lipoprotein cholesterol, TG—triglycerides. Rescaled [0, 1]
Identified patients with concurrent disease or medication. Some patients display starkly different profiles compared to their surroundings. Some of these (highlighted in blue) proved to be suffering from a concurrent disease or to be on another medication that influenced their profiles. However, this information was not disclosed within the data and was found independently, only to be later confirmed by the clinicians. This proves that our methodology can provide more understanding than would be obtainable from the traditional approach. Non-active patients are coloured green (group I) and yellow (group II). Active patients are coloured orange (group III) and red (group IV)
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request.
References
Abeysooriya M, Soria M, Kasu MS, Ziemann M (2021) Gene name errors: lessons not learned. PLoS Comput Biol 17(7):1008984
Aggarwal CC, Hinneburg A, Keim DA (2001) On the surprising behavior of distance metrics in high dimensional space. In: Database theory—ICDT 2001: 8th international conference London, UK, January 4–6, 2001 Proceedings 8. Springer, pp 420–434
Aletaha D, Nell VP, Stamm T, Uffmann M, Pflugbeil S, Machold K, Smolen JS (2005) Acute phase reactants add little to composite disease activity indices for rheumatoid arthritis: validation of a clinical activity score. Arthritis Res Ther 7(4):1–11
Alm E, Arkin AP (2003) Biological networks. Curr Opin Struct Biol 13(2):193–202
Arbelaitz O, Gurrutxaga I, Muguerza J, Pérez JM, Perona I (2013) An extensive comparative study of cluster validity indices. Pattern Recognit 46(1):243–256
Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. In: International AAAI Conference on Weblogs and Social Media. http://www.aaai.org/ocs/index.php/ICWSM/09/paper/view/154
Bel Mufti G, Bertrand P, El Moubarki L (2005) Determining the number of groups from measures of cluster stability. In: Proceedings of international symposium on applied stochastic models and data analysis, pp 404–412
Belkin M, Niyogi P (2003) Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput 15(6):1373–1396
Belkin M, Niyogi P, Sindhwani V (2006) Manifold regularization: a geometric framework for learning from labeled and unlabeled examples. J Mach Learn Res 7(85):2399–2434
Ben-Hur A, Guyon I (2003) Detecting stable clusters using principal component analysis. Methods Mol Biol 224:159–182. https://doi.org/10.1385/1-59259-364-X:159
Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E (2008) Fast unfolding of communities in large networks. J Stat Mech Theory Exp 2008(10):10008
Boers M, Tugwell P, Felson D, Van Riel P, Kirwan J, Edmonds J, Smolen J, Khaltaev N, Muirden K (1994) World health organization and international league of associations for rheumatology core endpoints for symptom modifying antirheumatic drugs in rheumatoid arthritis clinical trials. J Rheumatol Suppl 41:86–89
Budtarad N, Prawjaeng J, Leelahavarong P, Pilasant S, Chanjam C, Narongroeknawin P, Kitumnuaypong T, Katchamart W (2023) Efficacy and safety of biologic, biosimilars and targeted synthetic dmards in moderate-to-severe rheumatoid arthritis with inadequate response to methotrexate: a systematic review and network meta-analysis. medRxiv, 2023-01
Chakraborty A, Dutta T, Mondal S, Nath A (2018) Application of graph theory in social media. Int J Comput Sci Eng 6(10):722–729
Chen W-Y, Song Y, Bai H, Lin C-J, Chang EY (2010) Parallel spectral clustering in distributed systems. IEEE Trans Pattern Anal Mach Intell 33(3):568–586
Crowson CS, Gunderson TM, Davis III JM, Myasoedova E, Kronzer VL, Coffey CM, Atkinson EJ (2022) Using unsupervised machine learning methods to cluster comorbidities in a population-based cohort of patients with rheumatoid arthritis. Arthritis Care Res 75(2):210–219. https://doi.org/10.1002/acr.24973
Davies DL, Bouldin DW (1979) A cluster separation measure. IEEE Trans Pattern Anal Mach Intell 2:224–227
Dimitriadou E, Dolničar S, Weingessel A (2002) An examination of indexes for determining the number of clusters in binary data sets. Psychometrika 67(1):137–159
Dunn JC (1974) Well-separated clusters and optimal fuzzy partitions. J Cybern 4(1):95–104
Endres DM, Schindelin JE (2003) A new metric for probability distributions. IEEE Trans Inf Theory 49(7):1858–1860
Gallo J, Kriegova E, Kudelka M, Lostak J, Radvansky M (2020) Gender differences in contribution of smoking, low physical activity, and high BMI to increased risk of early reoperation after TKA. J Arthroplasty 35(6):1545–1557
Halkidi M, Batistakis Y, Vazirgiannis M (2002) Cluster validity methods: Part i. ACM SIGMOD Rec 31(2):40–45
Hamming R (1980) Entropy and Shannon’s first theorem. Coding and information theory, vol 107. Prentice-Hall, Englewood Cliffs
Hric D, Darst RK, Fortunato S (2014) Community detection in networks: structural communities versus ground truth. Phys Rev E 90(6):062805
Hubert L, Arabie P (1985) Comparing partitions. J Classif 2:193–218
Jaccard P (1901) Étude comparative de la distribution florale dans une portion des alpes et des jura. Bull Soc Vaudoise Sci Nat 37:547–579
Jaccard P (1912) The distribution of the flora in the alpine zone. 1. New Phytol 11(2):37–50
Jacomy M, Venturini T, Heymann S, Bastian M (2014) ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE 9(6):98679
Jia H, Ding S, Xu X, Nie R (2014) The latest research progress on spectral clustering. Neural Comput Appl 24:1477–1486
Jung SM, Park K-S, Kim K-J (2021) Clinical phenotype with high risk for initiation of biologic therapy in rheumatoid arthritis: a data-driven cluster analysis. Clin Exp Rheumatol 39(6):1282–1290
Kannan R, Vempala S, Vetta A (2004) On clusterings: good, bad and spectral. J ACM 51(3):497–515
Kotulska A, Kopeć-Mȩdrek M, Grosicka A, Kubicka M, Kucharz EJ (2015) Correlation between erythrocyte sedimentation rate and c-reactive protein level in patients with rheumatic diseases. Rheumatology 53(5):243–246
Landewé R, van der Heijde D, van der Linden S, Boers M (2006) Twenty-eight-joint counts invalidate the DAS28 remission definition owing to the omission of the lower extremity joints: a comparison with the original DAS remission. Ann Rheum Dis 65(5):637–641
Lee YC, Frits ML, Iannaccone CK, Weinblatt ME, Shadick NA, Williams DA, Cui J (2014) Subgrou** of patients with rheumatoid arthritis based on pain, fatigue, inflammation, and psychosocial factors. Arthritis Rheumatol 66(8):2006–2014
Leeb BF, Andel I, Sautner J, Bogdan M, Maktari A, Nothnagl T, Rintelen B (2005) Disease activity measurement of rheumatoid arthritis: Comparison of the simplified disease activity index (SDAI) and the disease activity score including 28 joints (DAS28) in daily routine. Arthritis Care Res 53(1):56–60
Lewis D (2021) Autocorrect errors in excel still creating genomics headache. Nature. https://www.nature.com/articles/d41586-021-02211-4
Ma C, Lv Q, Teng S, Yu Y, Niu K, Yi C (2017) Identifying key genes in rheumatoid arthritis by weighted gene co-expression network analysis. Int J Rheum Dis 20(8):971–979
Mahalanobis PC (1936) On the generalised distance in statistics. In: Proceedings of the National Institute of Science of India, vol 12, pp 49–55
Manukyan G, Papajik T, Mikulkova Z, Urbanova R, Kraiczova VS, Savara J, Kudelka M, Turcsanyi P, Kriegova E (2020) High CXCR3 on leukemic cells distinguishes IgHVmut from IgHVunmut in chronic lymphocytic leukemia: evidence from CD5high and CD5low clones. J Immunol Res 2020:7084268. https://doi.org/10.1155/2020/7084268
Matta J, Singh V, Auten T, Sanjel P (2023) Inferred networks, machine learning, and health data. PLoS ONE 18(1):0280910
Maulik U, Bandyopadhyay S (2002) Performance evaluation of some clustering algorithms and validity indices. IEEE Trans Pattern Anal Mach Intell 24(12):1650–1654
Mikulkova Z, Manukyan G, Turcsanyi P, Kudelka M, Urbanova R, Savara J, Ochodkova E, Brychtova Y, Molinsky J, Simkovic M (2021) Deciphering the complex circulating immune cell microenvironment in chronic lymphocytic leukaemia using patient similarity networks. Sci Rep 11(1):322
Nataliani Y, Yang M-S (2019) Powered gaussian kernel spectral clustering. Neural Comput Appl 31:557–572
Newman ME (2004) Detecting community structure in networks. Eur Phys J B 38:321–330
Newman ME, Girvan M (2004) Finding and evaluating community structure in networks. Phys Rev E 69(2):026113
Ng A, Jordan M, Weiss Y (2001) On spectral clustering: analysis and an algorithm. Advances in neural information processing systems, vol 14
Ochodkova E, Zehnalova S, Kudelka M (2017) Graph construction based on local representativeness. In: International computing and combinatorics conference. Springer, pp 654–665
Pascual D, Pla F, Sánchez JS (2010) Cluster validation using information stability measures. Pattern Recognit Lett 31(6):454–461
Pavlopoulos GA, Secrier M, Moschopoulos CN, Soldatos TG, Kossida S, Aerts J, Schneider R, Bagos PG (2011) Using graph theory to analyze biological networks. BioData Min 4:1–27
Pincus T, Morley S (2001) Cognitive-processing bias in chronic pain: a review and integration. Psychol Bull 127(5):599
Platzer A, Alasti F, Smolen JS, Aletaha D, Radner H, Blüml S (2022) Trajectory clusters of radiographic progression in patients with rheumatoid arthritis: associations with clinical variables. Ann Rheum Dis 81(2):175–183
Qiao L, Zhang L, Chen S, Shen D (2018) Data-driven graph construction and graph learning: a review. Neurocomputing 312:336–351
Riazzoli J, Nilsson J-Å, Teleman A, Petersson IF, Rantapää-Dahlqvist S, Jacobsson LTH, van Vollenhoven RF (2010) Patient-reported 28 swollen and tender joint counts accurately represent RA disease activity and can be used to assess therapy responses at the group level. Rheumatology 49(11):2098–2103
Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65
Saade A, Krzakala F, Zdeborová L (2014) Spectral clustering of graphs with the Bethe hessian. Advances in neural information processing systems, vol 27
Scott J (2000) Social network analysis: a handbook, 2nd edn
Silva TC, Zhao L (2012) Network-based high level data classification. IEEE Trans Neural Netw Learn Syst 23(6):954–970
Silva TC, Zhao L, Zequan L, Zhao Y, **n C (2018) Machine learning in complex networks, vol 1. Springer, Cham
Smolen JS, Breedveld FC, Burmester GR, Bykerk V, Dougados M, Emery P, Kvien TK, Navarro-Compán MV, Oliver S, Schoels M (2016) Treating rheumatoid arthritis to target: 2014 update of the recommendations of an international task force. Ann Rheum Dis 75(1):3–15
Smolen J, Breedveld F, Schiff M, Kalden J, Emery P, Eberl G, Van Riel P, Tugwell P (2003) A simplified disease activity index for rheumatoid arthritis for use in clinical practice. Rheumatology 42(2):244–257
Song X, Zhang Y, Dai E, Du H, Wang L (2019) Mechanism of action of celastrol against rheumatoid arthritis: a network pharmacology analysis. Int Immunopharmacol 74:105725
Spalenza MA, Pirovani JP, de Oliveira E (2021) Structures discovering for optimizing external clustering validation metrics. In: Intelligent systems design and applications: 19th international conference on intelligent systems design and applications (ISDA 2019) Held December 3-5, 2019 19. Springer, pp 150–161
Theodoridis S, Koutroumbas K (2009) Chapter 16 - cluster validity. In: Theodoridis S, Koutroumbas K (eds) Pattern recognition, 4th edn. Academic Press, Boston, pp 863–913
Tibshirani R, Walther G, Hastie T (2001) Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc Ser B (Stat Methodol) 63(2):411–423
Trajerova M, Kriegova E, Mikulkova Z, Savara J, Kudelka M, Gallo J (2022) Knee osteoarthritis phenotypes based on synovial fluid immune cells correlate with clinical outcome trajectories. Osteoarthr Cartil 30(12):1583–1592
Turchin A, Goldberg SI, Breydo E, Shubina M, Einbinder JS (2011) Copy/paste documentation of lifestyle counseling and glycemic control in patients with diabetes: true to form? Arch Intern Med 171(15):1393–1400
Turcsanyi P, Kriegova E, Kudelka M, Radvansky M, Kruzova L, Urbanova R, Schneiderova P, Urbankova H, Papajik T (2019) Improving risk-stratification of patients with chronic lymphocytic leukemia using multivariate patient similarity networks. Leuk Res 79:60–68
van der Heijde DM, Jacobs JW (1998) The original “DAS’’ and the “DAS28’’ are not interchangeable: comment on the articles by Prevoo et al. Arthritis Rheumatism 41(5):942–943
Van der Maaten L, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9(11):2579–2605
van Gestel AM, Haagsma CJ, van Riel PL (1998) Validation of rheumatoid arthritis improvement criteria that include simplified joint counts. Arthritis Rheumatism 41(10):1845–1850
Vieira VdF, Xavier CR, Evsukoff AG (2020) A comparative study of overlap** community detection methods from the perspective of the structural properties. Appl Netw Sci 5(1):1–42
Wang B, Mezlini AM, Demir F, Fiume M, Tu Z, Brudno M, Haibe-Kains B, Goldenberg A (2014) Similarity network fusion for aggregating data types on a genomic scale. Nat Methods 11(3):333–337
Wang M, Hua X-S, Tang J, Hong R (2009) Beyond distance measurement: constructing neighborhood similarity for video annotation. IEEE Trans Multimed 11(3):465–476
Wang YR, Bickel PJ (2017) Likelihood-based model selection for stochastic block models. Ann Statist 45(2):500–528. https://doi.org/10.1214/16-AOS1457
Wootton R (2001) Telemedicine. BMJ 323(7312):557–560
**ao J, Wang R, Cai X, Ye Z (2021) Coupling of co-expression network analysis and machine learning validation unearthed potential key genes involved in rheumatoid arthritis. Front Genet 12:604714
Xu Y, Olman V, Xu D (2002) Clustering gene expression data using a graph-theoretic approach: an application of minimum spanning trees. Bioinformatics 18(4):536–545
Yadalam PK, Sivasankari T, Rengaraj S, Mugri MH, Sayed M, Khan SS, Kamil MA, Bhandi S, Raj AT, Patil S (2022) Gene interaction network analysis reveals IFI44L as a drug target in rheumatoid arthritis and periodontitis. Molecules 27(9):2749
Yan X (2016) Bayesian model selection of stochastic block models. In: 2016 IEEE/ACM international conference on advances in social networks analysis and mining (ASONAM). IEEE, pp 323–328
Zehnalova S, Kudelka M, Platos J (2014a) Local representativeness in vector data. In: 2014 IEEE international conference on systems, man, and cybernetics (SMC). IEEE, pp 894–899
Zehnalova S, Kudelka M, Platos J, Horak Z (2014b) Local representatives in weighted networks. In: 2014 IEEE/ACM international conference on advances in social networks analysis and mining (ASONAM 2014), pp 870–875
Zhang L, Losin EAR, Ashar YK, Koban L, Wager TD (2021) Gender biases in estimation of others’ pain. J Pain 22(9):1048–1059
Zhang Y, Bai M, Zhang B, Liu C, Guo Q, Sun Y, Wang D, Wang C, Jiang Y, Lin N (2015) Uncovering pharmacological mechanisms of Wu-tou decoction acting on rheumatoid arthritis through systems approaches: drug-target prediction, network analysis and experimental validation. Sci Rep 5(1):9463
Zheng W, Rao S (2015) Knowledge-based analysis of genetic associations of rheumatoid arthritis to inform studies searching for pleiotropic genes: a literature review and network analysis. Arthritis Res Ther 17(1):1–9
Zhu J, Wang J, Dong Y, Song Y, Huang P (2023) Correlation between ultrasonographic scores and American college of rheumatology recommended rheumatoid arthritis disease activity measures: a systematic review and network meta-analysis. J Ultrasound 2023:1–9
Acknowledgements
The authors kindly thank all study participants.
Funding
The authors acknowledge the support of SGS, VSB-Technical University of Ostrava (Grant No. SP2023/076), NU20-06-00269, IGA_LF_2023_010, and MH_CZ-DRO (FNOL_00098892).
Author information
Authors and Affiliations
Contributions
OJ contributed to all sections and conducted the experimental part. EO contributed to all sections and the revision. MK designed the study and contributed to revision. EK contributed to the revision. PH and MS provided the data and contributed as domain experts. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the ethics committee of University Hospital Olomouc and Palacky University Olomouc.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Janca, O., Ochodkova, E., Kriegova, E. et al. Real-world data in rheumatoid arthritis: patient similarity networks as a tool for clinical evaluation of disease activity. Appl Netw Sci 8, 57 (2023). https://doi.org/10.1007/s41109-023-00582-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41109-023-00582-3