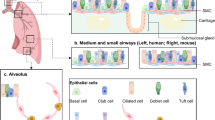
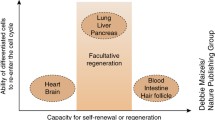
Abstract
Purpose of Review
Throughout the lifespan, lung injury impedes the primary critical function essential for life-respiration. To repair quickly and efficiently is critical and is orchestrated by a diverse repertoire of progenitor cells and their niche. This review incorporates knowledge gained from early studies in lung epithelial morphogenesis and cell fate and explores its relevance to more recent findings of lung progenitor and stem cells in development and regeneration.
Recent Findings
Cell fate in the lung is organized into an early specification phase and progressive differentiation phase in lung development. The advent of single-cell analysis combined with lineage analysis and projections is uncovering new functional cell types in the lung, providing a topographical atlas for progenitor cell lineage commitment during development, homeostasis, and regeneration.
Summary
Lineage commitment of lung progenitor cells is spatiotemporally regulated during development. Single-cell sequencing technologies have significantly advanced our understanding of the similarities and differences between developmental and regenerative cell fate trajectories. Subsequent unraveling of the molecular mechanisms underlying these cell fate decisions will be essential to manipulating progenitor cells for regeneration.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Franks TJ, Colby TV, Travis WD, Tuder RM, Reynolds HY, Brody AR, et al. Resident cellular components of the human lung: current knowledge and goals for research on cell phenoty** and function. Proc Am Thorac Soc. 2008;5(7):763–6. https://doi.org/10.1513/pats.200803-025HR.
Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(-/-) mouse embryos. Dev Biol. 1999;209(1):60–71. https://doi.org/10.1006/dbio.1999.9234.
Alanis DM, Chang DR, Akiyama H, Krasnow MA, Chen J. Two nested developmental waves demarcate a compartment boundary in the mouse lung. Nat Commun. 2014;5:3923. https://doi.org/10.1038/ncomms4923.
Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for Sox2 in the develo** and adult mouse trachea. Development. 2009;136(11):1899–907. https://doi.org/10.1242/dev.034629.
•• Plasschaert LW, Zilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560(7718):377–81. https://doi.org/10.1038/s41586-018-0394-6Identifies a new recently discovered cell type called pulmonary ionocyte in the conducting airways which is a major source of CFTR (gene mutated in cystic fibrosis) activity in the lungs being regulated by Notch and FoxI1 signaling.
•• Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560(7718):319–24. https://doi.org/10.1038/s41586-018-0393-7Defines the place of the ionocyte in the epithelial hierarchy by proving that the newly identified ionocyte cell type is a part of high turnover set of epithelial cells that are continuously replenished by basal progenitor cells regulated by FoxI1.
Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J, Cardoso WV. Notch signaling controls the balance of ciliated and secretory cell fates in develo** airways. Development. 2009;136(13):2297–307. https://doi.org/10.1242/dev.034884.
Morimoto M, Liu Z, Cheng HT, Winters N, Bader D, Kopan R. Canonical Notch signaling in the develo** lung is required for determination of arterial smooth muscle cells and selection of Clara versus ciliated cell fate. J Cell Sci. 2010;123(Pt 2):213–24. https://doi.org/10.1242/jcs.058669.
• Rawlins EL, Ostrowski LE, Randell SH, Hogan BL. Lung development and repair: contribution of the ciliated lineage. Proc Natl Acad Sci U S A. 2007;104(2):410–7. https://doi.org/10.1073/pnas.0610770104Describes elegant lineage tracing experiments using the Id2 Cre driver which established the progressive specification and differentiation of the distal tip of the airways.
Reynolds SD, Reynolds PR, Pryhuber GS, Finder JD, Stripp BR. Secretoglobins SCGB3A1 and SCGB3A2 define secretory cell subsets in mouse and human airways. Am J Respir Crit Care Med. 2002;166(11):1498–509. https://doi.org/10.1164/rccm.200204-285OC.
Post LC, Ternet M, Hogan BL. Notch/Delta expression in the develo** mouse lung. Mech Dev. 2000;98(1–2):95–8.
Ito T, Udaka N, Yazawa T, Okudela K, Hayashi H, Sudo T, et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. 2000;127(18):3913–21.
Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci U S A. 2002;99(16):10482–7. https://doi.org/10.1073/pnas.152238499.
Rawlins EL, Clark CP, Xue Y, Hogan BL. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136(22):3741–5. https://doi.org/10.1242/dev.037317.
• Frank DB, Penkala IJ, Zepp JA, Sivakumar A, Linares-Saldana R, Zacharias WJ, et al. Early lineage specification defines alveolar epithelial ontogeny in the murine lung. Proc Natl Acad Sci U S A. 2019. https://doi.org/10.1073/pnas.1813952116Suggests that alveolar specification at the distal tips initiated concurrent with the branching morphogenesis during the pseudoglandular stage changing the timeline of alveolar specification.
• Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–5. https://doi.org/10.1038/nature13173First study to use single-cell RNA sequencing to describe the lineage hierarchies to support the progressive specification and differentiation model of alveologenesis.
• Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507(7491):190–4. https://doi.org/10.1038/nature12930Proposes the existence of a novel bipotent progenitor population at the distal tip of the airways that gave rise to the alveolar type I and type II cells during alveologenesis and its relevance in cancer.
Steinhart Z, Angers S. Wnt signaling in development and tissue homeostasis. Development. 2018;145(11). https://doi.org/10.1242/dev.146589.
De Langhe SP, Reynolds SD. Wnt signaling in lung organogenesis. Organogenesis. 2008;4(2):100–8. https://doi.org/10.4161/org.4.2.5856.
Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, et al. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev Cell. 2009;17(2):290–8. https://doi.org/10.1016/j.devcel.2009.06.005.
Ostrin EJ, Little DR, Gerner-Mauro KN, Sumner EA, Ríos-Corzo R, Ambrosio E, et al. β-Catenin maintains lung epithelial progenitors after lung specification. Development. 2018;145(5):dev160788. https://doi.org/10.1242/dev.160788.
Harris-Johnson KS, Domyan ET, Vezina CM, Sun X. Beta-catenin promotes respiratory progenitor identity in mouse foregut. Proc Natl Acad Sci U S A. 2009;106(38):16287–92. https://doi.org/10.1073/pnas.0902274106.
De Langhe SP, Sala FG, Del Moral P-M, Fairbanks TJ, Yamada KM, Warburton D, et al. Dickkopf-1 (DKK1) reveals that fibronectin is a major target of Wnt signaling in branching morphogenesis of the mouse embryonic lung. Developmental Biology. 2005;277(2):316–31. https://doi.org/10.1016/j.ydbio.2004.09.023.
Shu W, Guttentag S, Wang Z, Andl T, Ballard P, Lu MM, et al. Wnt/beta-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev Biol. 2005;283(1):226–39. https://doi.org/10.1016/j.ydbio.2005.04.014.
Hashimoto S, Chen H, Que J, Brockway BL, Drake JA, Snyder JC, et al. beta-Catenin-SOX2 signaling regulates the fate of develo** airway epithelium. J Cell Sci. 2012;125(Pt 4):932–42. https://doi.org/10.1242/jcs.092734.
Okubo T, Hogan BL. Hyperactive Wnt signaling changes the developmental potential of embryonic lung endoderm. J Biol. 2004;3(3):11. https://doi.org/10.1186/jbiol3.
Frank DB, Peng T, Zepp JA, Snitow M, Vincent TL, Penkala IJ, et al. Emergence of a wave of Wnt signaling that regulates lung alveologenesis by controlling epithelial self-renewal and differentiation. Cell Reports. 2016;17(9):2312–25. https://doi.org/10.1016/j.celrep.2016.11.001.
Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4(3):215–66. https://doi.org/10.1002/wdev.176.
Serls AE, Doherty S, Parvatiyar P, Wells JM, Deutsch GH. Different thresholds of fibroblast growth factors pattern the ventral foregut into liver and lung. Development. 2005;132(1):35–47. https://doi.org/10.1242/dev.01570.
Yuan T, Volckaert T, Chanda D, Thannickal VJ, De Langhe SP. Fgf10 signaling in lung development, homeostasis, disease, and repair after injury. Front Genet. 2018;9:418. https://doi.org/10.3389/fgene.2018.00418.
Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, et al. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21(1):138–41. https://doi.org/10.1038/5096.
• El Agha E, Herold S, Al Alam D, Quantius J, MacKenzie B, Carraro G, et al. Fgf10-positive cells represent a progenitor cell population during lung development and postnatally. Development. 2014;141(2):296–306. https://doi.org/10.1242/dev.099747Shows the existence of two temporal waves of FGF signaling and its contribution to lung progenitor specification and development.
Volckaert T, Campbell A, Dill E, Li C, Minoo P, De Langhe S. Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors. Development. 2013;140(18):3731. https://doi.org/10.1242/dev.096560.
Abler LL, Mansour SL, Sun X. Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev Dyn. 2009;238(8):1999–2013. https://doi.org/10.1002/dvdy.22032.
Chang DR, Martinez Alanis D, Miller RK, Ji H, Akiyama H, McCrea PD, et al. Lung epithelial branching program antagonizes alveolar differentiation. Proc Natl Acad Sci U S A. 2013;110(45):18042–51. https://doi.org/10.1073/pnas.1311760110.
Li J, Wang Z, Chu Q, Jiang K, Li J, Tang N. The strength of mechanical forces determines the differentiation of alveolar epithelial cells. Developmental Cell. 2018;44(3):297–312.e5. https://doi.org/10.1016/j.devcel.2018.01.008.
Misra JR, Irvine KD. The Hippo signaling network and its biological functions. Annual Review of Genetics. 2018;52(1):65–87. https://doi.org/10.1146/annurev-genet-120417-031621.
Lin C, Yao E, Zhang K, Jiang X, Croll S, Thompson-Peer K, et al. YAP is essential for mechanical force production and epithelial cell proliferation during lung branching morphogenesis. eLife. 2017;6:e21130. https://doi.org/10.7554/eLife.21130.
Mahoney John E, Mori M, Szymaniak Aleksander D, Varelas X, Cardoso WV. The Hippo pathway effector Yap controls patterning and differentiation of airway epithelial progenitors. Developmental Cell. 2014;30(2):137–50. https://doi.org/10.1016/j.devcel.2014.06.003.
Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, et al. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. American Journal of Physiology-Renal Physiology. 2008;294(3):F542–F53. https://doi.org/10.1152/ajprenal.00201.2007.
Lange AW, Sridharan A, Xu Y, Stripp BR, Perl A-K, Whitsett JA. Hippo/Yap signaling controls epithelial progenitor cell proliferation and differentiation in the embryonic and adult lung. Journal of Molecular Cell Biology. 2014;7(1):35–47. https://doi.org/10.1093/jmcb/mju046%.
Lin C, Yao E, Chuang PT. A conserved MST1/2-YAP axis mediates Hippo signaling during lung growth. Dev Biol. 2015;403(1):101–13. https://doi.org/10.1016/j.ydbio.2015.04.014.
Nantie LB, Young RE, Paltzer WG, Zhang Y, Johnson RL, Verheyden JM, et al. Lats1/2 inactivation reveals Hippo function in alveolar type I cell differentiation during lung transition to air breathing. Development. 2018;145(21):dev163105. https://doi.org/10.1242/dev.163105.
Zhou B, Flodby P, Luo J, Castillo DR, Liu Y, Yu F-X, et al. Claudin-18–mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. The Journal of Clinical Investigation. 2018;128(3):970–84. https://doi.org/10.1172/JCI90429.
• van Soldt BJ, Qian J, Li J, Tang N, Lu J, Cardoso WV. Yap and its subcellular localization have distinct compartment-specific roles in the develo** lung. Development. 2019:dev.175810. https://doi.org/10.1242/dev.175810Consists of elegantly designed experiments that were able to resolve the controversies surrounding the complex role of HIPPO-YAP pathway in lung specification.
Weiss A, Attisano L. The TGFbeta superfamily signaling pathway. Wiley Interdisciplinary Reviews: Developmental Biology. 2013;2(1):47–63. https://doi.org/10.1002/wdev.86.
Aschner Y, Downey GP. Transforming growth factor-beta: master regulator of the respiratory system in health and disease. Am J Respir Cell Mol Biol. 2016;54(5):647–55. https://doi.org/10.1165/rcmb.2015-0391TR.
Domyan ET, Ferretti E, Throckmorton K, Mishina Y, Nicolis SK, Sun X. Signaling through BMP receptors promotes respiratory identity in the foregut via repression of Sox2. Development. 2011;138(5):971–81. https://doi.org/10.1242/dev.053694.
Li Y, Gordon J, Manley NR, Litingtung Y, Chiang C. Bmp4 is required for tracheal formation: a novel mouse model for tracheal agenesis. Dev Biol. 2008;322(1):145–55. https://doi.org/10.1016/j.ydbio.2008.07.021.
Que J, Choi M, Ziel JW, Klingensmith J, Hogan BL. Morphogenesis of the trachea and esophagus: current players and new roles for noggin and Bmps. Differentiation. 2006;74(7):422–37. https://doi.org/10.1111/j.1432-0436.2006.00096.x.
Weaver M, Yingling JM, Dunn NR, Bellusci S, Hogan BL. Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development. 1999;126(18):4005.
Sun J, Chen H, Chen C, Whitsett JA, Mishina Y, Bringas P Jr, et al. Prenatal lung epithelial cell-specific abrogation of Alk3-bone morphogenetic protein signaling causes neonatal respiratory distress by disrupting distal airway formation. The American Journal of Pathology. 2008;172(3):571–82. https://doi.org/10.2353/ajpath.2008.070286.
Eblaghie MC, Reedy M, Oliver T, Mishina Y, Hogan BL. Evidence that autocrine signaling through Bmpr1a regulates the proliferation, survival and morphogenetic behavior of distal lung epithelial cells. Dev Biol. 2006;291(1):67–82. https://doi.org/10.1016/j.ydbio.2005.12.006.
Kong Y, Glickman J, Subramaniam M, Shahsafaei A, Allamneni KP, Aster JC, et al. Functional diversity of notch family genes in fetal lung development. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2004;286(5):L1075–L83. https://doi.org/10.1152/ajplung.00438.2002.
Tsao PN, Chen F, Izvolsky KI, Walker J, Kukuruzinska MA, Lu J, et al. Gamma-secretase activation of notch signaling regulates the balance of proximal and distal fates in progenitor cells of the develo** lung. J Biol Chem. 2008;283(43):29532–44. https://doi.org/10.1074/jbc.M801565200.
Tsao P-N, Wei S-C, Wu M-F, Huang M-T, Lin H-Y, Lee M-C, et al. Notch signaling prevents mucous metaplasia in mouse conducting airways during postnatal development. Development. 2011;138(16):3533. https://doi.org/10.1242/dev.063727.
Zhang S, Loch AJ, Radtke F, Egan SE, Xu K. Jagged1 is the major regulator of notch-dependent cell fate in proximal airways. Developmental Dynamics. 2013;242(6):678–86. https://doi.org/10.1002/dvdy.23965.
Borges M, Linnoila RI, van de Velde HJ, Chen H, Nelkin BD, Mabry M, et al. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature. 1997;386(6627):852–5. https://doi.org/10.1038/386852a0.
Jia S, Wildner H, Birchmeier C. Insm1 controls the differentiation of pulmonary neuroendocrine cells by repressing Hes1. Dev Biol. 2015;408(1):90–8. https://doi.org/10.1016/j.ydbio.2015.10.009.
Morimoto M, Nishinakamura R, Saga Y, Kopan R. Different assemblies of Notch receptors coordinate the distribution of the major bronchial Clara, ciliated and neuroendocrine cells. Development. 2012;139(23):4365–73. https://doi.org/10.1242/dev.083840.
Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, et al. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development. 2009;136(10):1751. https://doi.org/10.1242/dev.029249.
Mori M, Mahoney JE, Stupnikov MR, Paez-Cortez JR, Szymaniak AD, Varelas X, et al. Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development. 2015;142(2):258. https://doi.org/10.1242/dev.116855.
Leach JP, Morrisey EE. Repairing the lungs one breath at a time: How dedicated or facultative are you? Genes Dev. 2018;32(23–24):1461–71. https://doi.org/10.1101/gad.319418.118.
Lynch TJ, Engelhardt JF. Progenitor cells in proximal airway epithelial development and regeneration. J Cell Biochem. 2014;115(10):1637–45. https://doi.org/10.1002/jcb.24834.
•• Tata A, Kobayashi Y, Chow RD, Tran J, Desai A, Massri AJ, et al. Myoepithelial cells of submucosal glands can function as reserve stem cells to regenerate airways after injury. Cell Stem Cell. 2018;22(5):668–83 e6. https://doi.org/10.1016/j.stem.2018.03.018Uncovers the submucosal myoepithelial cell as a progenitor cell in airway regeneration.
•• Lynch TJ, Anderson PJ, Rotti PG, Tyler SR, Crooke AK, Choi SH, et al. Submucosal gland myoepithelial cells are reserve stem cells that can regenerate mouse tracheal epithelium. Cell Stem Cell. 2018;22(5):653–67 e5. https://doi.org/10.1016/j.stem.2018.03.017Uncovers the submucosal myoepithelial cell as a progenitor cell in airway regeneration.
Boers JE, Ambergen AW, Thunnissen FB. Number and proliferation of basal and parabasal cells in normal human airway epithelium. Am J Respir Crit Care Med. 1998;157(6 Pt 1):2000–6. https://doi.org/10.1164/ajrccm.157.6.9707011.
Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106(31):12771–5. https://doi.org/10.1073/pnas.0906850106.
Yang Y, Riccio P, Schotsaert M, Mori M, Lu J, Lee DK, et al. Spatial-temporal lineage restrictions of embryonic p63(+) progenitors establish distinct stem cell pools in adult airways. Dev Cell. 2018;44(6):752–61 e4. https://doi.org/10.1016/j.devcel.2018.03.001.
Watson JK, Rulands S, Wilkinson AC, Wuidart A, Ousset M, Van Keymeulen A, et al. Clonal dynamics reveal two distinct populations of basal cells in slow-turnover airway epithelium. Cell Rep. 2015;12(1):90–101. https://doi.org/10.1016/j.celrep.2015.06.011.
Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503(7475):218–23. https://doi.org/10.1038/nature12777.
Zhao R, Fallon TR, Saladi SV, Pardo-Saganta A, Villoria J, Mou H, et al. Yap tunes airway epithelial size and architecture by regulating the identity, maintenance, and self-renewal of stem cells. Dev Cell. 2014;30(2):151–65. https://doi.org/10.1016/j.devcel.2014.06.004.
Balasooriya GI, Goschorska M, Piddini E, Rawlins EL. FGFR2 is required for airway basal cell self-renewal and terminal differentiation. Development. 2017;144(9):1600. https://doi.org/10.1242/dev.135681.
Ghosh M, Brechbuhl HM, Smith RW, Li B, Hicks DA, Titchner T, et al. Context-dependent differentiation of multipotential keratin 14-expressing tracheal basal cells. Am J Respir Cell Mol Biol. 2011;45(2):403–10. https://doi.org/10.1165/rcmb.2010-0283OC.
Pardo-Saganta A, Law BM, Tata PR, Villoria J, Saez B, Mou H, et al. Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell. 2015;16(2):184–97. https://doi.org/10.1016/j.stem.2015.01.002.
• Rock JR, Gao X, Xue Y, Randell SH, Kong YY, Hogan BL. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 2011;8(6):639–48. https://doi.org/10.1016/j.stem.2011.04.003Identifies NOTCH-mediated mechanisms that regulate differentiation of basal cells towards the secretory lineages during repair post lung injury.
• Pardo-Saganta A, Tata PR, Law BM, Saez B, Chow RD-W, Prabhu M, et al. Parent stem cells can serve as niches for their daughter cells. Nature. 2015;523:597. https://doi.org/10.1038/nature14553Reveals that airway basal stem/progenitor cells continuously supply a NOTCH ligand to their daughter secretory cells which was critical in maintaining the progenitor pool.
Kumar PA, Hu Y, Yamamoto Y, Hoe NB, Wei TS, Mu D, et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147(3):525–38. https://doi.org/10.1016/j.cell.2011.10.001.
Ray S, Chiba N, Yao C, Guan X, McConnell AM, Brockway B, et al. Rare SOX2(+) Airway progenitor cells generate KRT5(+) cells that repopulate damaged alveolar parenchyma following influenza virus infection. Stem Cell Reports. 2016;7(5):817–25. https://doi.org/10.1016/j.stemcr.2016.09.010.
• Vaughan AE, Brumwell AN, ** Y, Gotts JE, Brownfield DG, Treutlein B, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517(7536):621–5. https://doi.org/10.1038/nature14112Characterizes a rare lineage-negative epithelial stem/progenitor (LNEP) cells present within normal distal lung which assembled into the KRT5 pods that form during influenza injuries and importantly identified NOTCH-mediated pathways that promoted the differentiation of these cells to reconstitute the alveoli.
Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517(7536):616–20. https://doi.org/10.1038/nature13903.
Kanegai CM, ** Y, Donne ML, Gotts JE, Driver IH, Amidzic G, et al. Persistent pathology in influenza-infected mouse lungs. Am J Respir Cell Mol Biol. 2016;55(4):613–5. https://doi.org/10.1165/rcmb.2015-0387LE.
•• ** Y, Kim T, Brumwell AN, Driver IH, Wei Y, Tan V, et al. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nature Cell Biology. 2017;19:904. https://doi.org/10.1038/ncb3580Uncovers unique pathways critical for converting KRT5+ pods into alveolar epithelium for lung regeneration after influenza injury.
Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4(6):525–34. https://doi.org/10.1016/j.stem.2009.04.002.
Guha A, Vasconcelos M, Cai Y, Yoneda M, Hinds A, Qian J, et al. Neuroepithelial body microenvironment is a niche for a distinct subset of Clara-like precursors in the develo** airways. Proc Natl Acad Sci U S A. 2012;109(31):12592–7. https://doi.org/10.1073/pnas.1204710109.
Guha A, Deshpande A, Jain A, Sebastiani P, Cardoso WV. Uroplakin 3a(+) Cells are a distinctive population of epithelial progenitors that contribute to airway maintenance and post-injury repair. Cell Rep. 2017;19(2):246–54. https://doi.org/10.1016/j.celrep.2017.03.051.
Giangreco A, Reynolds SD, Stripp BR. Terminal bronchioles harbor a unique airway stem cell population that localizes to the bronchoalveolar duct junction. Am J Pathol. 2002;161(1):173–82. https://doi.org/10.1016/S0002-9440(10)64169-7.
Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121(6):823–35. https://doi.org/10.1016/j.cell.2005.03.032.
•• Liu Q, Liu K, Cui G, Huang X, Yao S, Guo W, et al. Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat Genet. 2019;51(4):728–38. https://doi.org/10.1038/s41588-019-0346-6Using a novel dual recombinase lineage tracing tool, confirms the existence of BASCs and their importance in lung regeneration after injury.
•• Salwig I, Spitznagel B, Vazquez-Armendariz AI, Khalooghi K, Guenther S, Herold S, et al. Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo. EMBO J. 2019. https://doi.org/10.15252/embj.2019102099Using a novel dual lineage tracing reconstitutive Cre recombinase tool, confirms the existence of BASCs and their importance in lung regeneration after injury.
Evans MJ, Cabral LJ, Stephens RJ, Freeman G. Renewal of alveolar epithelium in the rat following exposure to NO2. Am J Pathol. 1973;70(2):175–98.
•• Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123(7):3025–36. https://doi.org/10.1172/JCI68782First study to show the self-renewal and the differentiation capabilities of the AT2 cells in the alveoli.
Rock JR, Barkauskas CE, Cronce MJ, Xue Y, Harris JR, Liang J, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A. 2011;108(52):E1475–83. https://doi.org/10.1073/pnas.1117988108.
Jain R, Barkauskas CE, Takeda N, Bowie EJ, Aghajanian H, Wang Q, et al. Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat Commun. 2015;6:6727. https://doi.org/10.1038/ncomms7727.
Chung MI, Bujnis M, Barkauskas CE, Kobayashi Y, Hogan BLM. Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development. 2018;145(9). https://doi.org/10.1242/dev.163014.
LaCanna R, Liccardo D, Zhang P, Tragesser L, Wang Y, Cao T, et al. Yap/Taz regulate alveolar regeneration and resolution of lung inflammation. J Clin Invest. 2019;130:2107–22. https://doi.org/10.1172/JCI125014.
Finn J, Sottoriva K, Pajcini KV, Kitajewski JK, Chen C, Zhang W, et al. Dlk1-mediated temporal regulation of Notch signaling is required for differentiation of alveolar type II to type I cells during repair. Cell Rep. 2019;26(11):2942–54 e5. https://doi.org/10.1016/j.celrep.2019.02.046.
Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, et al. Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest. 2011;121(7):2855–62. https://doi.org/10.1172/JCI57673.
• Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018;359(6380):1118–23. https://doi.org/10.1126/science.aam6603Highlights the importance of Wnt signaling in regulating the stemness and transdifferential potential of a subset of AT2 cells and its role in mediating the cross talk with the neighboring Wnt secreting fibroblasts during homeostasis and injury.
• Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 2018;555:251. https://doi.org/10.1038/nature25786. Identifies a stable, Wnt-responsive subpopulation of AT2 cells that are poised for regeneration during homeostasis which upon influenza-mediated injury contributes significantly to the regeneration of alveoli.
Acknowledgments
Due to space limitations, we apologize to our scientific colleagues whose work could not be cited. We would like to thank Dr. Jarod Zepp for the critical review of this manuscript.
Funding
This work was supported by grants from the National Institutes of Health K08-HL140129 (D.B.F), the Parker B. Francis Foundation (D.B.F.), and the W.W. Smith Charitable Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Aravind Sivakumar and David B. Frank declare that they have no conflict of interest
Human and Animal Rights and Informed Consent
This manuscript does not contain any original studies involving human or animal subjects.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Prenatal Therapies
Rights and permissions
About this article
Cite this article
Sivakumar, A., Frank, D.B. Paradigms That Define Lung Epithelial Progenitor Cell Fate in Development and Regeneration. Curr Stem Cell Rep 5, 133–144 (2019). https://doi.org/10.1007/s40778-019-00166-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40778-019-00166-x