Abstract
Purpose of review
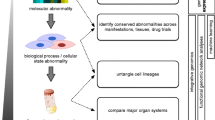
The emergence of genomic data science stands poised to revolutionize our molecular understanding of the heterogeneity of complex diseases including systemic autoimmune diseases. In systemic sclerosis (SSc), bulk and single-cell transcriptomics have provided a new lens into the heterogeneity of this complex condition, both in terms of molecular heterogeneity, treatment response, and cell types important for the disease.
Recent findings
Transcriptomics has revealed reproducible patterns of gene expression among SSc patients. These conserved patterns of gene expression provide insights into SSc etiology, and evidence suggests that these groups may have important implications for treatment decisions by targeting specific patients. Integration and analyses of publicly available data are providing new insights into the disease. Single-cell technologies are illuminating cell types that may be important in pathogenesis. The disease trajectory for SSc remains difficult to predict, but the interactions between adaptive and innate immune cells with tissue-resident stromal cells may play an important role.
Summary
The heterogeneity in SSc can be broken down and quantified using molecular methods that range from bulk analysis to single cells. Further study of cellular and molecular dynamics in end-target tissues is likely to result in better disease management through personalized, data-driven treatment decisions.

Similar content being viewed by others
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Peoples C, Medsger TA Jr, Lucas M, Rosario BL, Feghali-Bostwick CA. Gender differences in systemic sclerosis: relationship to clinical features, serologic status and outcomes. J Scleroderma Relat Disord. 2016;1(2):177–240. https://doi.org/10.5301/jsrd.5000209.
Allanore Y, Simms R, Distler O, Trojanowska M, Pope J, Denton CP, et al. Systemic sclerosis. Nat Rev Dis Primers. 2015;1:15002. https://doi.org/10.1038/nrdp.2015.2.
Hussein H, Lee P, Chau C, Johnson SR. The effect of male sex on survival in systemic sclerosis. J Rheumatol. 2014;41(11):2193–200. https://doi.org/10.3899/jrheum.140006.
Pérez-Bocanegra C, Solans-Laqué R, Simeón-Aznar CP, Campillo M, Fonollosa-Pla V, Vilardell-Tarrés M. Age-related survival and clinical features in systemic sclerosis patients older or younger than 65 at diagnosis. Rheumatology (Oxford). 2010;49(6):1112–7. https://doi.org/10.1093/rheumatology/keq046.
Barnes J, Mayes MD. Epidemiology of systemic sclerosis: incidence, prevalence, survival, risk factors, malignancy, and environmental triggers. Curr Opin Rheumatol. 2012;24(2):165–70. https://doi.org/10.1097/BOR.0b013e32834ff2e8.
Gelber AC, Manno RL, Shah AA, Woods A, Le EN, Boin F, et al. Race and association with disease manifestations and mortality in scleroderma: a 20-year experience at the Johns Hopkins Scleroderma Center and review of the literature. Medicine (Baltimore). 2013;92(4):191–205. https://doi.org/10.1097/MD.0b013e31829be125.
Blanco I, Mathai S, Shafiq M, Boyce D, Kolb TM, Chami H, et al. Severity of systemic sclerosis-associated pulmonary arterial hypertension in African Americans. Medicine (Baltimore). 2014;93(5):177–85. https://doi.org/10.1097/MD.0000000000000032.
Morgan ND, Shah AA, Mayes MD, Domsic RT, Medsger TA Jr, Steen VD, et al. Clinical and serological features of systemic sclerosis in a multicenter African American cohort: analysis of the genome research in African American scleroderma patients clinical database. Medicine (Baltimore). 2017;96(51):e8980. https://doi.org/10.1097/MD.0000000000008980.
Joseph CG, Darrah E, Shah AA, Skora AD, Casciola-Rosen LA, Wigley FM, et al. Association of the autoimmune disease scleroderma with an immunologic response to cancer. Science. 2014;343(6167):152–7. https://doi.org/10.1126/science.1246886.
Fanning GC, Welsh KI, Bunn C, Du Bois R, Black CM. HLA associations in three mutually exclusive autoantibody subgroups in UK systemic sclerosis patients. Br J Rheumatol. 1998;37(2):201–7.
Spencer-Green G, Alter D, Welch HG. Test performance in systemic sclerosis: anti-centromere and anti-Scl-70 antibodies. Am J Med. 1997;103(3):242–8.
Ho KT, Reveille JD. The clinical relevance of autoantibodies in scleroderma. Arthritis Res Ther. 2003;5(2):80–93.
Khimdas S, Harding S, Bonner A, Zummer B, Baron M, Pope J, et al. Associations with digital ulcers in a large cohort of systemic sclerosis: results from the Canadian Scleroderma Research Group registry. Arthritis Care Res. 2011;63(1):142–9. https://doi.org/10.1002/acr.20336.
Wirz EG, Jaeger VK, Allanore Y, Riemekasten G, Hachulla E, Distler O, et al. Incidence and predictors of cutaneous manifestations during the early course of systemic sclerosis: a 10-year longitudinal study from the EUSTAR database. Ann Rheum Dis. 2016;75(7):1285–92. https://doi.org/10.1136/annrheumdis-2015-207271.
Hamaguchi Y, Kodera M, Matsushita T, Hasegawa M, Inaba Y, Usuda T, et al. Clinical and immunologic predictors of scleroderma renal crisis in Japanese systemic sclerosis patients with anti-RNA polymerase III autoantibodies. Arthritis Rheum. 2015;67(4):1045–52. https://doi.org/10.1002/art.38994.
Steen VD, Medsger TA Jr. Severe organ involvement in systemic sclerosis with diffuse scleroderma. Arthritis Rheum. 2000;43(11):2437–44. https://doi.org/10.1002/1529-0131(200011)43:11<2437::AID-ANR10>3.0.CO;2-U.
LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA Jr, et al. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol. 1988;15(2):202–5.
Steen VD, Medsger TA Jr. Improvement in skin thickening in systemic sclerosis associated with improved survival. Arthritis Rheum. 2001;44(12):2828–35.
Shand L, Lunt M, Nihtyanova S, Hoseini M, Silman A, Black CM, et al. Relationship between change in skin score and disease outcome in diffuse cutaneous systemic sclerosis: application of a latent linear trajectory model. Arthritis Rheum. 2007;56(7):2422–31. https://doi.org/10.1002/art.22721.
Martyanov V, Whitfield ML. Molecular stratification and precision medicine in systemic sclerosis from genomic and proteomic data. Curr Opin Rheumatol. 2016;28(1):83–8. https://doi.org/10.1097/BOR.0000000000000237.
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–11. https://doi.org/10.1038/35000501.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. https://doi.org/10.1038/35021093.
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. https://doi.org/10.1073/pnas.191367098.
Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. https://doi.org/10.1038/nature11252.
Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–6. https://doi.org/10.1038/nm.3967.
Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. https://doi.org/10.1038/nature11404.
Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16(19):4864–75. https://doi.org/10.1158/1078-0432.CCR-10-0199.
Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. https://doi.org/10.1038/nature10166.
Verhaak RG, Wouters BJ, Erpelinck CA, Abbas S, Beverloo HB, Lugthart S, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94(1):131–4. https://doi.org/10.3324/haematol.13299.
Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74. https://doi.org/10.1056/NEJMoa1301689.
Hennigs A, Riedel F, Gondos A, Sinn P, Schirmacher P, Marme F, et al. Prognosis of breast cancer molecular subtypes in routine clinical care: a large prospective cohort study. BMC Cancer. 2016;16(1):734. https://doi.org/10.1186/s12885-016-2766-3.
Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer. 2011;11:143. https://doi.org/10.1186/1471-2407-11-143.
Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11(16):5678–85. https://doi.org/10.1158/1078-0432.CCR-04-2421.
Milano A, Pendergrass SA, Sargent JL, George LK, McCalmont TH, Connolly MK, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS One. 2008;3(7):e2696. https://doi.org/10.1371/journal.pone.0002696.
Assassi S, Wu M, Tan FK, Chang J, Graham TA, Furst DE, et al. Skin gene expression correlates of severity of interstitial lung disease in systemic sclerosis. Arthritis Rheum. 2013;65(11):2917–27. https://doi.org/10.1002/art.38101.
Hsu E, Shi H, Jordan RM, Lyons-Weiler J, Pilewski JM, Feghali-Bostwick CA. Lung tissues in patients with systemic sclerosis have gene expression patterns unique to pulmonary fibrosis and pulmonary hypertension. Arthritis Rheum. 2011;63(3):783–94. https://doi.org/10.1002/art.30159.
Vreća M, Zeković A, Damjanov N, Andjelković M, Ugrin M, Pavlović S, et al. Expression of TLR7, TLR9, JAK2, and STAT3 genes in peripheral blood mononuclear cells from patients with systemic sclerosis. J Appl Genet. 2018;59(1):59–66. https://doi.org/10.1007/s13353-017-0415-4.
Tan FK, Zhou X, Mayes MD, Gourh P, Guo X, Marcum C, et al. Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology (Oxford). 2006;45(6):694–702. https://doi.org/10.1093/rheumatology/kei244.
Beretta L, Barturen G, Vigone B, Bellocchi C, Hunzelmann N, De Langhe E, et al. Genome-wide whole blood transcriptome profiling in a large European cohort of systemic sclerosis patients. Ann Rheum Dis. 2020;79(9):1218–26. https://doi.org/10.1136/annrheumdis-2020-217116.
Taroni JN, Martyanov V, Huang CC, Mahoney JM, Hirano I, Shetuni B, et al. Molecular characterization of systemic sclerosis esophageal pathology identifies inflammatory and proliferative signatures. Arthritis Res Ther. 2015;17:194. https://doi.org/10.1186/s13075-015-0695-1.
Pendergrass SA, Hayes E, Farina G, Lemaire R, Farber HW, Whitfield ML, et al. Limited systemic sclerosis patients with pulmonary arterial hypertension show biomarkers of inflammation and vascular injury. PLoS One. 2010;5(8):e12106. https://doi.org/10.1371/journal.pone.0012106.
Pendergrass SA, Lemaire R, Francis IP, Mahoney JM, Lafyatis R, Whitfield ML. Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol. 2012;132(5):1363–73. https://doi.org/10.1038/jid.2011.472.
Hinchcliff M, Huang CC, Wood TA, Matthew Mahoney J, Martyanov V, Bhattacharyya S, et al. Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis. J Invest Dermatol. 2013;133(8):1979–89. https://doi.org/10.1038/jid.2013.130.
Taroni JN, Martyanov V, Mahoney JM, Whitfield ML. A functional genomic meta-analysis of clinical trials in systemic sclerosis: toward precision medicine and combination therapy. J Invest Dermatol. 2017;137(5):1033–41. https://doi.org/10.1016/j.jid.2016.12.007.
Assassi S, Swindell WR, Wu M, Tan FD, Khanna D, Furst DE, et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis Rheum. 2015;67(11):3016–26. https://doi.org/10.1002/art.39289.
• Moon SJ, Bae JM, Park KS, Tagkopoulos I, Kim KJ. Compendium of skin molecular signatures identifies key pathological features associated with fibrosis in systemic sclerosis. Ann Rheum Dis. 2019;78(6):817–25. https://doi.org/10.1136/annrheumdis-2018-214778. Performed an integrative analysis of eight gene expression datasets, including 173 SSc patients, to identify four unique clusters of patients and activity signatures for SSc-relevent pathways.
•• Xu X, Ramanujam M, Visvanathan S, Assassi S, Liu Z, Li L. Transcriptional insights into pathogenesis of cutaneous systemic sclerosis using pathway driven meta-analysis assisted by machine learning methods. PLoS One. 2020;15(11):e0242863. https://doi.org/10.1371/journal.pone.0242863. Computational analysis of nine geneexpression datasets to identify 80 gene expression signatures to further stratify the original intrinsic subsets into eight unique subtypes.
Mahoney JM, Taroni J, Martyanov V, Wood TA, Greene CS, Pioli PA, et al. Systems level analysis of systemic sclerosis shows a network of immune and profibrotic pathways connected with genetic polymorphisms. PLoS Comput Biol. 2015;11(1):e1004005. https://doi.org/10.1371/journal.pcbi.1004005.
Steen VD, Medsger TA. Changes in causes of death in systemic sclerosis, 1972-2002. Ann Rheum Dis. 2007;66(7):940–4. https://doi.org/10.1136/ard.2006.066068.
Allanore Y, Matucci-Cerinic M, Distler O. Treatment of systemic sclerosis: is there any hope for the future? RMD Open. 2016;2(2):e000260. https://doi.org/10.1136/rmdopen-2016-000260.
Matucci-Cerinic M, Steen VD, Furst DE, Seibold JR. Clinical trials in systemic sclerosis: lessons learned and outcomes. Arthritis Res Ther. 2007;9(Suppl 2):S7. https://doi.org/10.1186/ar2191.
Gordon JK, Martyanov V, Magro C, Wildman HF, Wood TA, Huang WT, et al. Nilotinib (Tasigna) in the treatment of early diffuse systemic sclerosis: an open-label, pilot clinical trial. Arthritis Res Ther. 2015;17:213. https://doi.org/10.1186/s13075-015-0721-3.
Rice LM, Padilla CM, McLaughlin SR, Mathes A, Ziemek J, Goummih S, et al. Fresolimumab treatment decreases biomarkers and improves clinical symptoms in systemic sclerosis patients. J Clin Invest. 2015;125(7):2795–807. https://doi.org/10.1172/JCI77958.
Stifano G, Sornasse T, Rice LM, Na L, Chen-Harris H, Khanna D, et al. Skin gene expression is prognostic for the trajectory of skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheum. 2018;70(6):912–9. https://doi.org/10.1002/art.40455.
Sargent JL, Milano A, Bhattacharyya S, Varga J, Connolly MK, Chang HY, et al. A TGFbeta-responsive gene signature is associated with a subset of diffuse scleroderma with increased disease severity. J Invest Dermatol. 2010;130(3):694–705. https://doi.org/10.1038/jid.2009.318.
Hinchcliff M, Toledo DM, Taroni JN, Wood TA, Franks JM, Ball MS, et al. Mycophenolate mofetil treatment of systemic sclerosis reduces myeloid cell numbers and attenuates the inflammatory gene signature in skin. J Invest Dermatol. 2018;138(6):1301–10. https://doi.org/10.1016/j.jid.2018.01.006.
Baker KP, Edwards BM, Main SH, Choi GH, Wager RE, Halpern WG, et al. Generation and characterization of LymphoStat-B, a human monoclonal antibody that antagonizes the bioactivities of B lymphocyte stimulator. Arthritis Rheum. 2003;48(11):3253–65. https://doi.org/10.1002/art.11299.
•• Spiera R, Hummers L, Chung L, Frech TM, Domsic R, Hsu V, et al. Safety and efficacy of lenabasum in a phase II, randomized, placebo-controlled trial in adults with systemic sclerosis. Arthritis Rheum. 2020;72(8):1350–60. https://doi.org/10.1002/art.41294. Gene expression analysis of inflammation and fibrosis gene signatures in particioants of a double-blind, placebo-controlled trial of lenabasum.
Chakravarty EF, Martyanov V, Fiorentino D, Wood TA, Haddon DJ, Jarrell JA, et al. Gene expression changes reflect clinical response in a placebo-controlled randomized trial of abatacept in patients with diffuse cutaneous systemic sclerosis. Arthritis Res Ther. 2015;17:159. https://doi.org/10.1186/s13075-015-0669-3.
•• Khanna D, Spino C, Johnson S, Chung L, Whitfield ML, Denton CP, et al. Abatacept in early diffuse cutaneous systemic sclerosis: results of a phase II investigator-initiated, multicenter, double-blind, randomized, placebo-controlled trial. Arthritis Rheum. 2020;72(1):125–36. https://doi.org/10.1002/art.41055. Blinded gene expression analysis of participants in the Abatacept to Treat Diffuse Cutaneous Systemic Sclerosis (ASSET) trial demonstrating that intrinsic gene expression subsets responded significantly differently to treatment.
Medsger TA Jr. Natural history of systemic sclerosis and the assessment of disease activity, severity, functional status, and psychologic well-being. Rheum Dis Clin N Am. 2003;29(2):255–73 vi.
Shah AA, Wigley FM. My approach to the treatment of scleroderma. Mayo Clin Proc. 2013;88(4):377–93. https://doi.org/10.1016/j.mayocp.2013.01.018.
Sullivan KM, Goldmuntz EA, Keyes-Elstein L, McSweeney PA, Pinckney A, Welch B, et al. Myeloablative autologous stem-cell transplantation for severe scleroderma. N Engl J Med. 2018;378(1):35–47.
•• Franks JM, Martyanov V, Wang Y, Wood TA, Pinckney A, Crofford LJ, et al. Machine learning predicts stem cell transplant response in severe scleroderma. Ann Rheum Dis. 2020;79(12):1608–15. https://doi.org/10.1136/annrheumdis-2020-217033. Gene expression analysis of peripheral blood from participantys in the Scleroderma Cyclophosphamide or Transplantation (SCOT) trial demonstrating prognostic potential on intrinsic molecular subsets.
Gordon JK, Martyanov V, Franks JM, Bernstein EJ, Szymonifka J, Magro C, et al. Belimumab for the treatment of early diffuse systemic sclerosis: results of a randomized, double-blind, placebo-controlled, pilot trial. Arthritis Rheum. 2018;70(2):308–16. https://doi.org/10.1002/art.40358.
Altman N, Krzywinski M. The curse(s) of dimensionality. Nat Methods. 2018;15(6):399–400. https://doi.org/10.1038/s41592-018-0019-x.
Zimek A, Schuber E, Kriegel H. A survey on unsupervised outlier detection in high-dimensional numerical data. Statistical Analysis and Data Mining. 2012;5(5):363–87.
Krzywinski M, Altman N. Multiple linear regression. Nat Methods. 2015;12(12):1103–4.
Reality check on reproducibility. Nature. 2016;533(7604):437. doi: 10.1038/533437a.
Franks JM, Cai G, Whitfield ML. Feature specific quantile normalization enables cross-platform classification of molecular subtypes using gene expression data. Bioinformatics. 2018. https://doi.org/10.1093/bioinformatics/bty026.
Taroni JN, Greene CS, Martyanov V, Wood TA, Christmann RB, Farber HW, et al. A novel multi-network approach reveals tissue-specific cellular modulators of fibrosis in systemic sclerosis. Genome Med. 2017;9(1):27. https://doi.org/10.1186/s13073-017-0417-1.
Wang Y, Franks JM, Yang M, Toledo DM, Wood TA, Hinchcliff M, et al. Regulator combinations identify systemic sclerosis patients with more severe disease. JCI Insight. 2020;5(17). https://doi.org/10.1172/jci.insight.137567.
• Karimizadeh E, Sharifi-Zarchi A, Nikaein H, Salehi S, Salamatian B, Elmi N, et al. Analysis of gene expression profiles and protein-protein interaction networks in multiple tissues of systemic sclerosis. BMC Med Genet. 2019;12(1):199. https://doi.org/10.1186/s12920-019-0632-2. Integrative analysis of ten published gene expressiondatasets across multiple tissues to identify conserved disease signatures.
•• Kim KJ, Moon SJ, Park KS. Network-based modeling of drug effects on disease module in systemic sclerosis. Sci Rep. 2020;10(1):13393. https://doi.org/10.1038/s41598-020-70280-y. Computational network analysis to identify drug targets and potential therapeutics from 179 unique SSc-associated genes.
Franks JM, Martyanov V, Cai G, Wang Y, Li Z, Wood TA, et al. A machine learning classifier for assigning individual patients with systemic sclerosis to intrinsic molecular subsets. Arthritis Rheum. 2019. https://doi.org/10.1002/art.40898.
Lofgren S, Hinchcliff M, Carns M, Wood T, Aren K, Arroyo E, et al. Integrated, multicohort analysis of systemic sclerosis identifies robust transcriptional signature of disease severity. JCI Insight. 2016;1(21):e89073. Epub 2016/12/22. https://doi.org/10.1172/jci.insight.89073.
• Showalter K, Spiera R, Magro C, Agius P, Martyanov V, Franks JM, et al. Machine learning integration of scleroderma histology and gene expression identifies fibroblast polarisation as a hallmark of clinical severity and improvement. Ann Rheum Dis. 2021;80(2):228–37. https://doi.org/10.1136/annrheumdis-2020-217840. Gene expression and histology were performed on participants from two clinical trials. Machine learning algorithms were used to link important signatures between the two data types and identify an activated fibroblast signature in patients who clinically improved.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. https://doi.org/10.1186/1471-2105-9-559.
• Skaug B, Khanna D, Swindell WR, Hinchcliff ME, Frech TM, Steen VD, et al. Global skin gene expression analysis of early diffuse cutaneous systemic sclerosis shows a prominent innate and adaptive inflammatory profile. Ann Rheum Dis. 2020;79(3):379–86. https://doi.org/10.1136/annrheumdis-2019-215894. RNA-sequencing performed on participants of the Prospective Registry for Early Systemic Sclerosis (PRESS) cohort demonstrating that adaptive and immune cell signatures are evident in early dcSSc and correlate to clinical measures.
Ha R, Mutasa S, Karcich J, Gupta N, Pascual Van Sant E, Nemer J, et al. Predicting breast cancer molecular subtype with MRI dataset utilizing convolutional neural network algorithm. J Digit Imaging. 2019. https://doi.org/10.1007/s10278-019-00179-2.
Young JD, Cai C, Lu X. Unsupervised deep learning reveals prognostically relevant subtypes of glioblastoma. BMC Bioinformatics. 2017;18(Suppl 11):381. https://doi.org/10.1186/s12859-017-1798-2.
Way GP, Greene CS. Extracting a biologically relevant latent space from cancer transcriptomes with variational autoencoders. Pac Symp Biocomput. 2018;23:80–91.
Apostolidis SA, Stifano G, Tabib T, Rice LM, Morse CM, Kahaleh B, et al. Single cell RNA sequencing identifies HSPG2 and APLNR as markers of endothelial cell injury in systemic sclerosis skin. Front Immunol. 2018;9:2191. https://doi.org/10.3389/fimmu.2018.02191.
Gaydosik AM, Tabib T, Domsic R, Khanna D, Lafyatis R, Fuschiotti P. Single-cell transcriptome analysis identifies skin-specific T-cell responses in systemic sclerosis. Ann Rheum Dis. 2021. https://doi.org/10.1136/annrheumdis-2021-220209.
Valenzi E, Bulik M, Tabib T, Morse C, Sembrat J, Trejo Bittar H, et al. Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Ann Rheum Dis. 2019;78(10):1379–87. https://doi.org/10.1136/annrheumdis-2018-214865.
• Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199(12):1517–36. https://doi.org/10.1164/rccm.201712-2410OC. Single-cell sequencing of lung explants from SSc patients and other clinical cases of pulmonary fibrosis to characterize cell types including a profibrotic alveolar macrophage which was only identified in patiens with fibrosis.
Moreno-Moral A, Bagnati M, Koturan S, Ko JH, Fonseca C, Harmston N, et al. Changes in macrophage transcriptome associate with systemic sclerosis and mediate. Ann Rheum Dis. 2018;77(4):596–601. https://doi.org/10.1136/annrheumdis-2017-212454.
Bhandari R, Ball MS, Martyanov V, Popovich D, Schaafsma E, Han S, et al. Profibrotic activation of human macrophages in systemic sclerosis. Arthritis Rheum. 2020;72(7):1160–9. https://doi.org/10.1002/art.41243.
•• Kobayashi S, Nagafuchi Y, Okubo M, Sugimori Y, Shirai H, Hatano H, et al. Integrated bulk and single-cell RNA-sequencing identified disease-relevant monocytes and a gene network module underlying systemic sclerosis. J Autoimmun. 2021;116:102547. https://doi.org/10.1016/j.jaut.2020.102547. Bulk RNA-sequencing and flow cytometry was performed on peripheral blood samples from 21 Japanese SSc patients and 13 matched controls to identify an inflammatory gene module. The authors leveraged publicly available single-cell data to identify a cluster of monocytes that may be linked to SSc pathophysiology.
Valenzi E, Tabib T, Papazoglou A, Sembrat J, Trejo Bittar HE, Rojas M, et al. Disparate interferon signaling and shared aberrant basaloid cells in single-cell profiling of idiopathic pulmonary fibrosis and systemic sclerosis-associated interstitial lung disease. Front Immunol. 2021;12:595811. https://doi.org/10.3389/fimmu.2021.595811.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Conflict of Interest
Jennifer Franks declares that she has no conflict of interest. Michael Whitfield reports grants, personal fees, and other from Celdara Medical LLC, grants and personal fees from Bristol Myers Squibb, grants and personal fees from Corbus Pharmaceuticals, personal fees from Abbvie, and personal fees from Acceleron, outside the submitted work.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Scleroderma
Rights and permissions
About this article
Cite this article
Franks, J.M., Whitfield, M.L. Insights Into Systemic Sclerosis from Gene Expression Profiling. Curr Treat Options in Rheum 7, 208–221 (2021). https://doi.org/10.1007/s40674-021-00183-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40674-021-00183-0