Abstract
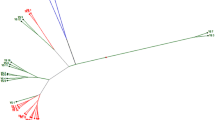
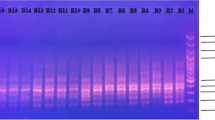
The genetic diversity and relationship was studied in a collection of 65 Brassica accessions, which included 54 B. rapa and 11 of five other Brassica species used as references. These 54 B. rapa accessions included 42 Chinese accessions and 12 exotic accessions. All accessions were analyzed by using 36 simple sequence repeats (SSR) and 43 sequence-related amplified polymorphism (SRAP) primers, and 401 polymorphic fragments were detected by SSR and SRAP markers. The average number of polymorphic fragments detected by SRAP markers was 6.23 ranging from 2 to 11 and that revealed by SSR was 3.69 ranging from 2 to 7. The unweighted pair-group method with arithmetic mean cluster analysis indicated that all accessions could be divided into five major clusters except three accessions as outliers. The exotic B. rapa accessions appeared in Cluster I except one yellow sarson accession from India, Chinese B. rapa revealed in Cluster II, and 11 accessions of other Brassica expressed them Cluster III, Cluster IV, and Cluster V. The results of principal component analysis and population structure analysis were in accordance with the cluster analysis. Molecular variance analysis revealed that the genetic variation was 26.10% among populations and 73.90% within Brassica species, which indicated existence of considerable genetic variation among exotic and Chinese B. rapa species, and exotic B. rapa can be used for broadening the genetic background of Chinese B. napus, and vice versa.



Similar content being viewed by others
References
Allender CJ, King GJ (2010) Origins of the amphidiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol 10:54
Annisa CS, Cowling WA (2013) Global genetic diversity in oilseed Brassica rapa. Crop Pasture Sci 64:993–1007
Chen S, Zou J, Cowling WA, Meng J (2010) Allelic diversity in a novel gene pool of canola-quality Brassica napus enriched with alleles from B. rapa and B carinata. Crop Pasture Sci 61:483–492
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fu YB, Gugel RK (2009) Genetic variability of Canadian elite cultivars of summer turnip rape Brassica rapa revealed by simple sequence repeat markers. Can J Plant Sci 89:865–874
Gómez-Campo C, Prakash S (1999) Origin and domestication. In: Gómez-Campo C (ed) Developments in plant genetics and breeding: biology of Brassica coeno species. Elsevier, Burlington, pp 33–58
Guo YM, Chen S, Li ZY, Cowling WA (2014) Center of origin and centers of diversity in an ancient crop, Brassica rapa (Turnip Rape). J Hered 105:555–565
He YT, Tu JX, Fu TD, Li DR, Chen BY (2002) Genetic diversity of germplasm resources of (Brassica campestris L.) in China by RAPD markers. Acta Agron Sin 28:693–703
Jesske T, Olberg B, Schierholt A, Becker HC (2013) Resynthesized lines from domesticated and wild Brassica taxa and their hybrids with B. napus L. genetic diversity and hybrid yield. Theor Appl Genet 126:1053–1065. doi:10.1007/s00122-012-2036-y
Li Q, Mei J, Zhang Y, Li J, Ge X, Li Z, Qian W (2013) A large-scale introgression of genomic components of Brassica rapa into B. napus by the bridge of hexaploid derived from hybridization between B. napus and B. oleracea. Theor Appl Genet 126:2073–2080. doi:10.1007/s00122-013-2119-4
Liu HL (1984) Origin and evolution of rapeseeds. Acta Agron Sin 10:9–18
Liu RH, Meng JL (2006) RFLP and AFLP analysis of inter and intra-specific variation of Brassica rapa and B. napus shows that B. rapa is an important genetic resource for B. napus improvement. Acta Gene Sin 33:814–823
McGrath JM, Quiros CE (1992) Genetic diversity at isozyme and RFLP loci in Brassica campestris as related to crop type and geographical origin. Theor Appl Genet 83:783–790
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4325
Ofori A, Becker HC, Kopisch-Obuch FJ (2008) Effect of crop improvement on genetic diversity in oilseed Brassica rapa (turnip-rape) cultivars, detected by SSR markers. J Appl Genet 49:207–212
Pino Del Carpio D, Basnet R, De Vos R, Maliepaard C, Visser R, Bonnema G (2011) The patterns of population differentiation in a Brassica rapa core collection. Theor Appl Genet 122:1105–1118
Prakash S, Hinata K (1980) Taxonomy, cytogenetics and origin of crop Brassicas, a review. Opera Bot 55:1–57
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Qian W, Meng J, Li M, Frauen M, Sass O, Noack J, Jung C (2006) Introgression of genomic components from Chinese Brassica rapa contributes to widening the genetic diversity in rapeseed (B. napus L.) with emphasis on the evolution of Chinese rapeseed. Theor Appl Genet 113:49–54
Ramchiary N, Lim YP (2011) Genetics of Brassica rapa L. In: Schmidt R, Bancroft I (eds) Genetics and genomics of the Brassicaceae. Springer, New York, pp 215–260
Reiner H, Holzner W, Ebermann R (1995) The development of turnip-type and oilseed-type Brassica rapa crops from the wild type in Europe-an overview of botanical, historical and linguistic facts. In: Rapeseed today and tomorrow, 9th International Rapeseed Congress, Cambridge, UK, pp 1066–1069
Rohlf FJ (1998) Numerical taxonomy and multivariate analysis system. Version 2.0. Exeter Software, Setauket, NY
Sokal RR, Michener CD (1958) A statistical method for evaluating systematic relationships. Univ Kans Sci Bull 38:1409–1438
Song KM, Osborn TC, Williams PH (1988) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs): 2. preliminary analysis of subspecies within B. rapa syn. campestris and B. oleracea. Theor Appl Genet 76:593–600
Song K, Osborn TC, Williams PH (1990) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs) 3. Genome relationships in Brassica and related genera and the origin of B. oleracea and B. rapa (syn. Campestris). Theor Appl Genet 79:497–506
Takahashi Y, Yokoi S, Takahata Y (2016) Genetic divergence of turnip (Brassica rapa L. em. Metzg. subsp. rapa) inferred from simple sequence repeats in chloroplast and nuclear genomes and morphology. Genetic Resour Crop Evol 63:869–879
Takuno S, Kawakara T, Ohnishi O (2007) Phylogenetic relationships among cultivated types of Brassica rapa L. em. Metzg.as revealed by AFLP analysis. Genet Resour Crop Evol 54:279–285
Tanhuanpää P, Erkkilä M, Tenhola-Roininen T, Tanskanen J, Manninen O (2016) SNP diversity within and among Brassica rapa accessions reveals no geographic differentiation. Genome 59:11–21. doi:10.1139/gen-2015-0118
U N (1935) Genomic analysis of Brassica with special reference to the experimental formation of B. napus and its peculiar mode of fertilization. Jpn J Bot 7:389–452
Warwick SI, James T, Falk KC (2008) AFLP-based molecular characterization of Brassica rapa and diversity in Canadian spring turnip rape cultivars. Plant Gene Resour 6:11–21
Zamani-Nour S, Clemens R, Möllers C (2013) Cytoplasmic diversity of Brassica napus L., Brassica oleracea L. and Brassica rapa L. as determined by chloroplast microsatellite markers. Genet Resour Crop Evol 60:953–965
Zhang RJ, Hu SW, Yan JQ, Sun GL (2013) Cytoplasmic diversity in Brassica rapa L. investigated by mitochondrial markers. Genet Resour Crop Evol 60:967–974
Zhao JY, Becker HC (1998) Genetic variation in Chinese and European oilseed rapa (B. napus) and Turnip rapa (B. campestris) analysed with isozymes. Acta Agron Sin 24:213–220
Zhao JJ, Wang XW, Deng B, Lou P, Wu J, Sun RF, Xu ZY, Vromans J, Koornneef M, Bonnema G (2005) Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor Appl Genet 110:1301–1314. doi:10.1007/s00122-005-1967-y
Zhao JJ, Paulo MJ, Jamar D, Lou P, Eeuwijk FV, Bonnema G, Vreugdenhil D, Koornneef M (2007) Association map** of leaf traits, flowering time, and phytate content in Brassica rapa. Genome 50:963–973
Zhao YG, Ofori A, Lu CM (2009) Genetic diversity of European and Chinese oilseed Brassica rapa cultivars from different breeding periods. Agric Sci China 8:31–938
Acknowledgements
This work was supported by the earmarked fund for China Agriculture Research System (CARS-13), National Key Technology R&D Program (2010BAD01B02), and a grant of Northwest A&F University for S.W. Hu.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Information of 43 pairs of SRAP primers (DOCX 23 kb)
Table S2
Information of 36 pairs of SSR primers (DOCX 21 kb)
Table S3
Scoring of a representative gel of SSR with primer BrGMS126 and accession no. in Table S3 correspond to Table 1 (XLS 547 kb)
Rights and permissions
About this article
Cite this article
Zhang, X., Chen, H., Channa, S.A. et al. Genetic diversity in Chinese and exotic Brassica rapa L. accessions revealed by SSR and SRAP markers. Braz. J. Bot 40, 973–982 (2017). https://doi.org/10.1007/s40415-017-0392-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-017-0392-1