Abstract
Myofibrillar myopathy (MFM) is a group of inherited muscular disorders characterized by myofibril dissolution and abnormal accumulation of degradation products. The diagnosis of muscular disorders based on clinical presentation is difficult due to phenotypic heterogeneity and overlap** symptoms. In addition, precise diagnosis does not always explain the disease etiopathology or the highly variable clinical course even among patients diagnosed with the same type of myopathy. The advent of high-throughput next-generation sequencing (NGS) has provided a successful and cost-effective strategy for identification of novel causative genes in myopathies, including MFM. So far, pathogenic mutations associated with MFM phenotype, including atypical MFM-like cases, have been identified in 17 genes: DES, CRYAB, MYOT, ZASP, FLNC, BAG3, FHL1, TTN, DNAJB6, PLEC, LMNA, ACTA1, HSPB8, KY, PYROXD1, and SQSTM + TIA1 (digenic). Most of these genes are also associated with other forms of muscle diseases. In addition, in many MFM patients, numerous genomic variants in muscle-related genes have been identified. The various myopathies and muscular dystrophies seem to form a single disease continuum; therefore, gene identification in one disease impacts the genetic etiology of the others. In this review, we describe the heterogeneity of the MFM genetic background focusing on the role of rare variants, the importance of whole genome sequencing in the identification of novel disease-associated mutations, and the emerging concept of variant load as the basis of the phenotypic heterogeneity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myofibrillar myopathy (MFM) is a clinically and genetically heterogeneous group of hereditary muscle diseases characterized by ectopic protein aggregates and a distinct pattern of myofibrillar disorganization. The disintegration of the myofibrils commences in the immediate proximity of the Z-disc and results in Z-disc streaming. This is followed by abnormal accumulation of degraded filamentous material in various patterns in the myofibril-free fiber regions around nuclei and under the sarcolemma. The abnormally accumulated proteins include desmin, myotilin, α-B-crystallin, filamin C, Bag3, actin, plectin, dystrophin, sarcoglycans, neural cell adhesion molecule (NCAM), gelsolin, ubiquitin, syncoilin, synemin, ** muscles, and proper muscle and heart performance. Development 142:994–1005. https://doi.org/10.1242/dev.115352 " href="/article/10.1007/s13353-018-0463-4#ref-CR67" id="ref-link-section-d34606543e1347">2015).
In addition, proteins encoded by BAG3, DNAJB6, and also HSPB8 are involved in chaperone-assisted selective autophagy (CASA). The CASA complex facilitates degradation of damaged Z-disc and is required to prevent protein aggregation (Arimura et al. 2011; Ulbricht et al. 2015). Also, sequestosome 1, itself not a chaperone, plays a role in the autophagy of ubiquitinated inclusions and thus prevents accumulation of abnormally folded proteins.
A recently described PYROXD1 myopathy (O’Grady et al. 2016), despite being initially associated with the Z-disc in co-localization studies, demonstrates that altered cellular redox regulation could be another pathomechanism in myopathies.
Overall, in the majority of MFM patients, the molecular dysfunction is associated with proteins involved in forming the structure and/or providing homeostasis of the Z-disc in the muscle fiber. It is anticipated that also potential novel genes involved in MFM yet to be identified might encode structural or chaperone proteins. Particular attention should be given to genes encoding proteins known to interact with the known MFM-associated proteins.
Potential non-monogenic inheritance
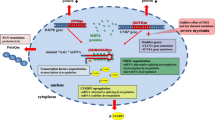
In about 50% of the MFM patients, the causative gene defect remains unknown. This might be partially due to the monogenic-centered concept of inherited diseases relying on Sanger sequencing of one or a few candidate genes at a time. However, di- and oligogenic heterozygosity has already been proposed as a potential disease mechanism for some metabolic and neurological diseases (Vockley et al. 2000; Schaaf et al. 2011). In some patients co-morbid, incomplete defects in multiple proteins of the same or of several independent pathways may lead to a disease, even though no causal mutation has been identified so far (Fig. 1). Such cases could be explained by the “epistatic effect” when one genetic variant is modulated by another one giving either a synergistic or an antagonistic effect on the phenotype (Phillips 2008). Another explanation is “synergistic heterozygosity,” when variants in two or more genes do not have a modulatory effect on each other but can cause partial defects in the same or intersecting pathways. In myopathies, several benign variants may accumulate in genes encoding proteins involved in a specific pathway (i.e., protein folding quality control) or in a single subcellular structure (such as the Z-disc or extrasarcomeric cytoskeleton). The cumulative effect of these variants, if exceeding a certain threshold, could affect muscle functioning and result in pathology such as MFM. We believe that such a cumulative effect of seemingly benign variants (with small effect when present in isolation) could explain the genetic background of a phenomenon observed in some families with children displaying a more severe disease phenotype than the one of their parents (Li et al. 2017). In addition, common polymorphisms might be disease course-modifying factors, such as LTPB4 variant in dystrophinopathy (Flanigan et al. 2013). We have observed a similar case in one MFM family, in which initial Sanger sequencing revealed a causative Q348P mutation in desmin co-segregating with the disease phenotype (Fichna et al. 2014). Subsequent whole exome sequencing revealed an additional S206L DNAJB6 mutation only in one patient with disease onset two decades earlier than of his mother’s, who also presented with a milder course of the disease (Fichna et al., manuscript in preparation/data not shown).
Pictorial comparison and difference between possible inheritance modes. a Typical monogenic inheritance with a pathogenic causal mutation in one gene. b Digenic inheritance, where mutations affecting the function of two genes are needed to produce the phenotype. c Oligogenic inheritance, where a number of mutations cause minor defects that together result in pathology. d Rare variant load, where accumulation of seemingly benign variants, including both mutations and polymorphisms, can lead to disease. In c and d, variants cause partial defects either in one pathway or in intersecting pathways, which together produce a cumulative effect leading to the disease. Circle symbolizes a gene; Asterisk stands for mutation/genetic variant
On the other hand, typical digenic inheritance has been demonstrated in facioscapulohumeral muscular dystrophy (Lemmers et al. 2012), in congenital myasthenic syndrome (Lam et al. 2017), and in calpainopathy (Sáenz and López de Munain 2017). Digenic inheritance has also been described in MFM-like cases caused by mutations in SQSTM1 and TIA1 (Niu et al. 2018).
New techniques and novel challenges
The strictly Mendelian concept of inheritance, with one mutation causing one clinical phenotype, is an oversimplification, as it was already elegantly presented on the example of phenylketonuria, the classical recessive disease (Scriver and Waters 1999). It was then proposed that modeling the interaction of a limited number of genes and understanding the molecular consequences of such interactions are a prerequisite for understanding genomic basis of phenotypic complexity of Mendelian disorders (Badano and Katsanis 2002). Nowadays, with the development and widespread application of various next-generation sequencing methods, it has become clear that not only a single gene can be associated with a specific disease (locus heterogeneity), but different variants in one gene can cause different phenotypes (allelic heterogeneity), and some variants can even be associated with multiple clinical phenotypes (Keith et al. 2014). The availability of exome and whole genome data for various conditions has challenged the classical definition of genetic causality and the concept of strictly monogenic disorders, providing evidence for the roots of heterogeneity and complexity of the human genome (Katsanis 2016). Many diseases with overlap** phenotypes have multiple genetic associations. Interestingly, to date, there have been over 400 genes associated with various neuromuscular disorders, with 407 associated only with “myopathy” in the OMIM database (omim.org).
Myopathies and muscular dystrophies seem to form a disease continuum; thus, the causative gene identification in one disease impacts the genetic etiology of the others. It could be hypothesized that complex patterns of inheritance, including oligogenic inheritance, account for a sizable fraction of myopathy cases, particularly the atypical ones, overlap** with other muscular disease forms. Specific genetic background, in particular comprising the genes for elements of pathways or structures involved in the pathogenesis of MFM, although difficult to pinpoint, may likely influence the phenotype. Modifying variants and even co-causal mutations may also explain the observed inter- and intrafamilial variability between patients harboring the same main causal mutation (Fichna et al. 2014). In MFM patients with an unknown genetic defect, the pathogenic and co-causative mutations (including copy number variants), as well as the modifying variants, could be located in non-coding, e.g., regulatory or deep intronic regions, not resolved by the whole exome sequencing approach, which highlights the superiority of the whole genome sequencing approach (Itan and Casanova 2015).
The concept of oligogenic inheritance should influence not only the approach to gene identification, but also genetic testing and counseling. NGS results are too complex to be easily interpreted by the health care professionals, who also struggle to pass comprehensive information on to the patients (Niemiec et al. 2018). However, one of the biggest challenges remains the discrimination between possible co-causative or modifying variants and the thousands of insignificant variants present in any genome. Advanced bioinformatic assessment of identified variants and comparison of worldwide NGS results offer some solution of this conundrum (Chakravorty and Hegde 2018).
A recent comprehensive genetic analysis using whole exome sequencing in a group of patients with various muscle disorders has shown that each patient bears over 30 rare (minor allele frequency, MAF < 1%) genetic variants potentially influencing the structure of relevant proteins and putatively related to the myopathic phenotype (Fichna et al. 2018). Interestingly, more of those rare variants were shared between patients with MFM and limb-girdle muscular dystrophy (LGMD) than between LGMD or MFM and a control group (Fichna et al., unpublished results). The hypothesis-driven approach used in the above study could have resulted in missing variants in genes not yet related to any muscle phenotype. Therefore, it was augmented by applying an additional filtering pipeline relating to muscle physiology or structure. It was aimed at the identification of very-rare variants (MAF < 0.1%) in genes expressed in the muscle and in genes coding for proteins from a broad interactome of muscle disease-related proteins. Using this combined approach, we were able to detect numerous variants often in genes coding for proteins involved in sarcomere structure and assembly, signal transduction, protein’s glycosylation, and folding or/and previously implicated in diverse muscle disorder (Fichna et al. 2018). Among these variants, potentially compromising protein structure and/or function could be novel pathogenic or phenotype-modifying ones. Therefore, we proposed that those variants could be part of a “variant burden,” contributing to deterioration of muscle cell molecular homeostasis and facilitating or modifying effects of the well-established causative MFM mutations. Moreover, mutations in genes encoding chaperones (CRYAB, BAG3, HSPB8, and DNAJB6) may likely unmask the burden of variants with minor effect. Chaperones rescue proteins misfolded by environmental stresses, but could also stabilize mutated proteins (Tomala and Korona 2008). Even if many MFM cases can be easily attributed clinically to mutations in a single gene, the high number of variants associated with myopathy and sometimes with specific phenotype features suggests that the variant load may be important even in patients with a well-defined primary pathogenic cause (Fichna et al. 2018). It is likely that in MFM-like cases with mutations in genes typically associated with other disorders (like ACTA1, HSPB8, LMNA, or SQSTM1), it is the variant load that ultimately determines the features of myofibrillar pathology.
The superiority of high-scale bioinformatic analysis over focused genetic studies lies in the possibility of repeating the analysis and applying novel knowledge, updated databases, better algorithms, and prediction tools. Therefore, genetic testing of MFM should be combined not only with bioinformatic analyses and deep phenoty**, but also with comprehensive analyses of transcripts and protein isoforms to pinpoint novel causal, co-causal, and modifying variants (Hennekam and Biesecker 2012). Identification of the modifying variants requires high-throughput analyses of combined genomic and clinical data on large groups of ethnically diverse patients with various muscle diseases and should be followed by functional studies. Such an approach will enable examination of groups of genes encoding entire pathways and cellular modules (McCarthy and MacArthur 2017). Better understanding of the importance of the modifying variants will inevitably transform the traditional descriptive classification of muscle diseases into a systemic and pathway-based view of clinical phenotypes (Thompson and Straub 2016).
Conclusions
There is mounting evidence that MFM and other myopathies should be viewed as oligogenic disorders in which variable clinical presentation can result from the combined effect of mutations in many genes. In this view, the inter- and intrafamilial variability could reflect a specific genetic background and the presence of sets of phenotype-modifying, co-causal mutations (variant burden). The concept of non-monogenic inheritance should influence not only the current approach to gene identification, but also genetic testing and counseling.
References
Arimura T, Ishikawa T, Nunoda S et al (2011) Dilated cardiomyopathy-associated BAG3 mutations impair Z-disc assembly and enhance sensitivity to apoptosis in cardiomyocytes. Hum Mutat 32:1481–1491. https://doi.org/10.1002/humu.21603
Badano JL, Katsanis N (2002) Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet 3:779–789. https://doi.org/10.1038/nrg910
Baker J, Riley G, Romero MR et al (2010) Identification of a Z-band associated protein complex involving KY, FLNC and IGFN1. Exp Cell Res 316:1856–1870. https://doi.org/10.1016/j.yexcr.2010.02.027
Beatham J, Romero R, Townsend SKM et al (2004) Filamin C interacts with the muscular dystrophy KY protein and is abnormally distributed in mouse KY deficient muscle fibres. Hum Mol Genet 13:2863–2874. https://doi.org/10.1093/hmg/ddh308
Blanco G, Coulton GR, Biggin A et al (2001) The kyphoscoliosis (ky) mouse is deficient in hypertrophic responses and is caused by a mutation in a novel muscle-specific protein. Hum Mol Genet 10:9–16
Castle JC, Zhang C, Shah JK et al (2008) Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 40:1416–1425. https://doi.org/10.1038/ng.264
Celik C, Uysal H, Heper AO, Karaoglan B (2005) Epidermolysis bullosa simplex associated with muscular dystrophy and cardiac involvement. J Clin Neuromuscul Dis 6:157–161. https://doi.org/10.1097/01.cnd.0000159779.32828.e7
Chakravorty S, Hegde M (2018) Inferring the effect of genomic variation in the new era of genomics. Hum Mutat 39:756–773. https://doi.org/10.1002/humu.23427
Dhawan PS, Liewluck T, Knapik J, Milone M (2018) Myofibrillar myopathy due to dominant LMNA mutations: a report of 2 cases. Muscle Nerve 57:E124–E126. https://doi.org/10.1002/mus.26036
Ehler E, Gautel M (2008) The sarcomere and Sarcomerogenesis. In: The sarcomere and skeletal muscle disease. Springer, New York, pp 1–14
Fichna JP, Karolczak J, Potulska-Chromik A, et al (2014) Two desmin gene mutations associated with myofibrillar myopathies in Polish families. PLoS One 9:e115470. doi: https://doi.org/10.1371/journal.pone.0115470
Fichna JP, Macias A, Piechota M, et al (2018) Whole-exome sequencing identifies novel pathogenic mutations and putative phenotype-influencing variants in Polish limb-girdle muscular dystrophy patients. Hum Genomics 12:34. doi: https://doi.org/10.1186/s40246-018-0167-1
Flanigan KM, Ceco E, Lamar K-M et al (2013) LTBP4 genotype predicts age of ambulatory loss in Duchenne muscular dystrophy. Ann Neurol 73:481–488. https://doi.org/10.1002/ana.23819
Förch P, Puig O, Martínez C et al (2002) The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5’ splice sites. EMBO J 21:6882–6892
Franzini-Armstrong C, Porter KR (1963) The Z disc of skeletal muscle fibrils. Zeitschrift für Zellforschung 61:661–672. https://doi.org/10.1007/BF00342617
Ghaoui R, Palmio J, Brewer J et al (2016) Mutations in HSPB8 causing a new phenotype of distal myopathy and motor neuropathy. Neurology 86:391–398. https://doi.org/10.1212/WNL.0000000000002324
Gonzaga-Jauregui C, Harel T, Gambin T et al (2015) Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep 12:1169–1183. https://doi.org/10.1016/j.celrep.2015.07.023
Graziano S, Kreienkamp R, Coll-Bonfill N, Gonzalo S (2018) Causes and consequences of genomic instability in laminopathies: replication stress and interferon response. Nucleus 9:258–275. https://doi.org/10.1080/19491034.2018.1454168
Hennekam RCM, Biesecker LG (2012) Next-generation sequencing demands next-generation phenoty**. Hum Mutat 33:884–886. https://doi.org/10.1002/humu.22048
Itan Y, Casanova J-L (2015) Can the impact of human genetic variations be predicted? Proc Natl Acad Sci U S A 112:11426–11427. https://doi.org/10.1073/pnas.1515057112
Izumi R, Niihori T, Aoki Y et al (2013) Exome sequencing identifies a novel TTN mutation in a family with hereditary myopathy with early respiratory failure. J Hum Genet 58:259–266. https://doi.org/10.1038/jhg.2013.9
Katsanis N (2016) The continuum of causality in human genetic disorders. Genome Biol 17:233. https://doi.org/10.1186/s13059-016-1107-9
Keith BP, Robertson DL, Hentges KE (2014) Locus heterogeneity disease genes encode proteins with high interconnectivity in the human protein interaction network. Front Genet 5:434. https://doi.org/10.3389/fgene.2014.00434
Klaavuniemi T, Ylänne J (2006) Zasp/Cypher internal ZM-motif containing fragments are sufficient to co-localize with alpha-actinin--analysis of patient mutations. Exp Cell Res 312:1299–1311. https://doi.org/10.1016/j.yexcr.2005.12.036
Knöll R, Buyandelger B, Lab M (2011) The sarcomeric Z-disc and Z-discopathies. J Biomed Biotechnol 2011:569628. https://doi.org/10.1155/2011/569628
Konieczny P, Fuchs P, Reipert S et al (2008) Myofiber integrity depends on desmin network targeting to Z-disks and costameres via distinct plectin isoforms. J Cell Biol 181:667–681. https://doi.org/10.1083/jcb.200711058
Krohne G, Benavente R (1986) The nuclear lamins. A multigene family of proteins in evolution and differentiation. Exp Cell Res 162:1–10
Lam C-W, Wong K-S, Leung H-W, Law C-Y (2017) Limb girdle myasthenia with digenic RAPSN and a novel disease gene AK9 mutations. Eur J Hum Genet 25:192–199. https://doi.org/10.1038/ejhg.2016.162
Lemmers RJLF, Tawil R, Petek LM et al (2012) Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet 44:1370–1374. https://doi.org/10.1038/ng.2454
Li F, **ao H, Hu Z et al (2018) Exploring the multifaceted roles of heat shock protein B8 (HSPB8) in diseases. Eur J Cell Biol 97:216–229. https://doi.org/10.1016/j.ejcb.2018.03.003
Li L, Bainbridge MN, Tan Y et al (2017) A potential oligogenic etiology of hypertrophic cardiomyopathy: a classic single-gene disorder. Circ Res 120:1084–1090. https://doi.org/10.1161/CIRCRESAHA.116.310559
Lin X, Ruiz J, Bajraktari I et al (2014) Z-disc-associated, alternatively spliced, PDZ motif-containing protein (ZASP) mutations in the actin-binding domain cause disruption of skeletal muscle actin filaments in myofibrillar myopathy. J Biol Chem 289:13615–13626. https://doi.org/10.1074/jbc.M114.550418
Matsumoto Y, Hayashi T, Inagaki N et al (2005) Functional analysis of titin/connectin N2-B mutations found in cardiomyopathy. J Muscle Res Cell Motil 26:367–374. https://doi.org/10.1007/s10974-005-9018-5
McCarthy MI, MacArthur DG (2017) Human disease genomics: from variants to biology. Genome Biol 18:20. https://doi.org/10.1186/s13059-017-1160-z
Niemiec E, Vears DF, Borry P, Howard HC (2018) Readability of informed consent forms for whole-exome and whole-genome sequencing. J Community Genet 9:143–151. https://doi.org/10.1007/s12687-017-0324-6
Niu Z, Pontifex CS, Berini S et al (2018) Myopathy with SQSTM1 and TIA1 variants: clinical and pathological features. Front Neurol 9:147. https://doi.org/10.3389/fneur.2018.00147
O’Grady GL, Best HA, Sztal TE et al (2016) Variants in the oxidoreductase PYROXD1 cause early-onset myopathy with internalized nuclei and myofibrillar disorganization. Am J Hum Genet 99:1086–1105. https://doi.org/10.1016/j.ajhg.2016.09.005
Ohlsson M, Hedberg C, Brådvik B et al (2012) Hereditary myopathy with early respiratory failure associated with a mutation in A-band titin. Brain 135:1682–1694. https://doi.org/10.1093/brain/aws103
Pfeffer G, Barresi R, Wilson IJ et al (2014) Titin founder mutation is a common cause of myofibrillar myopathy with early respiratory failure. J Neurol Neurosurg Psychiatry 85:331–338. https://doi.org/10.1136/jnnp-2012-304728
Phillips PC (2008) Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet 9:855–867. https://doi.org/10.1038/nrg2452
Raskin A, Lange S, Banares K et al (2012) A novel mechanism involving four-and-a-half LIM domain protein-1 and extracellular signal-regulated kinase-2 regulates titin phosphorylation and mechanics. J Biol Chem 287:29273–29284. https://doi.org/10.1074/jbc.M112.372839
Sáenz A, López de Munain A (2017) Dominant LGMD2A: alternative diagnosis or hidden digenism? Brain 140:e7. https://doi.org/10.1093/brain/aww281
Salmikangas P, van der Ven PFM, Lalowski M et al (2003) Myotilin, the limb-girdle muscular dystrophy 1A (LGMD1A) protein, cross-links actin filaments and controls sarcomere assembly. Hum Mol Genet 12:189–203
Sandell S, Huovinen S, Palmio J et al (2016) Diagnostically important muscle pathology in DNAJB6 mutated LGMD1D. Acta Neuropathol Commun 4:9. https://doi.org/10.1186/s40478-016-0276-9
Sarparanta J, Jonson PH, Golzio C et al (2012) Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet 44(450–455):S1–S2. https://doi.org/10.1038/ng.1103
Schaaf CP, Sabo A, Sakai Y et al (2011) Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum Mol Genet 20:3366–3375. https://doi.org/10.1093/hmg/ddr243
Schessl J, Taratuto AL, Sewry C et al (2009) Clinical, histological and genetic characterization of reducing body myopathy caused by mutations in FHL1. Brain 132:452–464. https://doi.org/10.1093/brain/awn325
Schoser B, Goebel HH, Janisch I et al (2009) Consequences of mutations within the C terminus of the FHL1 gene. Neurology 73:543–551. https://doi.org/10.1212/WNL.0b013e3181b2a4b3
Schröder R, Schoser B (2009) Myofibrillar myopathies: a clinical and myopathological guide. Brain Pathol 19:483–492. https://doi.org/10.1111/j.1750-3639.2009.00289.x
Scriver CR, Waters PJ (1999) Monogenic traits are not simple: lessons from phenylketonuria. Trends Genet 15:267–272
Selcen D (2011) Myofibrillar myopathies. Neuromuscul Disord 21:161–171. https://doi.org/10.1016/j.nmd.2010.12.007
Selcen D (2015) Severe congenital actin related myopathy with myofibrillar myopathy features. Neuromuscul Disord 25:488–492. https://doi.org/10.1016/j.nmd.2015.04.002
Selcen D, Bromberg MB, Chin SS, Engel AG (2011) Reducing bodies and myofibrillar myopathy features in FHL1 muscular dystrophy. Neurology 77:1951–1959. https://doi.org/10.1212/WNL.0b013e31823a0ebe
Selcen D, Engel AG (1993) Myofibrillar myopathy. In: Pagon RA, Adam MP, Ardinger HH, et al. (eds) GeneReviews(®). University of Washington, Seattle, Seattle (WA)
Selcen D, Engel AG (2004) Mutations in myotilin cause myofibrillar myopathy. Neurology 62:1363–1371
Selcen D, Muntoni F, Burton BK et al (2009) Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann Neurol 65:83–89. https://doi.org/10.1002/ana.21553
Shatunov A, Olivé M, Odgerel Z et al (2009) In-frame deletion in the seventh immunoglobulin-like repeat of filamin C in a family with myofibrillar myopathy. Eur J Hum Genet 17:656–663. https://doi.org/10.1038/ejhg.2008.226
Straussberg R, Schottmann G, Sadeh M et al (2016) Kyphoscoliosis peptidase (KY) mutation causes a novel congenital myopathy with core targetoid defects. Acta Neuropathol 132:475–478. https://doi.org/10.1007/s00401-016-1602-9
Thompson R, Straub V (2016) Limb-girdle muscular dystrophies - international collaborations for translational research. Nat Rev Neurol 12:294–309. https://doi.org/10.1038/nrneurol.2016.35
Thompson TG, Chan YM, Hack AA et al (2000) Filamin 2 (FLN2): a muscle-specific sarcoglycan interacting protein. J Cell Biol 148:115–126
Tomala K, Korona R (2008) Molecular chaperones and selection against mutations. Biol Direct 3:5. https://doi.org/10.1186/1745-6150-3-5
Ulbricht A, Gehlert S, Leciejewski B et al (2015) Induction and adaptation of chaperone-assisted selective autophagy CASA in response to resistance exercise in human skeletal muscle. Autophagy 11:538–546. https://doi.org/10.1080/15548627.2015.1017186
van Spaendonck-Zwarts KY, van Hessem L, Jongbloed JDH et al (2011) Desmin-related myopathy. Clin Genet 80:354–366. https://doi.org/10.1111/j.1399-0004.2010.01512.x
Vicart P, Caron A, Guicheney P et al (1998) A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet 20:92–95. https://doi.org/10.1038/1765
Vockley J, Rinaldo P, Bennett MJ et al (2000) Synergistic heterozygosity: disease resulting from multiple partial defects in one or more metabolic pathways. Mol Genet Metab 71:10–18. https://doi.org/10.1006/mgme.2000.3066
Winter L, Türk M, Harter PN et al (2016) Downstream effects of plectin mutations in epidermolysis bullosa simplex with muscular dystrophy. Acta Neuropathol Commun 4:44. https://doi.org/10.1186/s40478-016-0314-7
Wójtowicz I, Jabłońska J, Zmojdzian M et al (2015) Drosophila small heat shock protein CryAB ensures structural integrity of develo** muscles, and proper muscle and heart performance. Development 142:994–1005. https://doi.org/10.1242/dev.115352
Funding
The study on myofibrillar myopathy was funded by Polish National Science Centre grant (NCN 2012/05/D/NZ4/02978).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Communicated by: Michal Witt
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Fichna, J.P., Maruszak, A. & Żekanowski, C. Myofibrillar myopathy in the genomic context. J Appl Genetics 59, 431–439 (2018). https://doi.org/10.1007/s13353-018-0463-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-018-0463-4