Abstract
Feline parvovirus causes infectious diseases, and Chaphamaparvovirus is a novel type of feline parvovirus. The present study aims to establish a method that can be used in clinical rapid detection of feline Chaphamaparvovirus (FeChPV), for facilitate the timely and effective diagnosis and treatment of sick animals and shorten the diagnosis time of clinical diseases. The experimental samples in this study are from 20 cats undergoing physical examination in Hefei **n’an Animal Hospital. An SYBR Green I-based qPCR assay was performed to detect FeChPV. A pair of specific primers was designed based on the VP1 gene to perform the assay. The detection assay showed high sensitivity with a detection limit of 1.07 × 101 copies/μL and high specificity for detection of only the target virus. The coefficients of Ct value variation were calculated to assess the reproducibility of the qPCR assay, and the inter- and intra-assay ranged from 0.21 to 0.67% and 0.10 to 0.56%, respectively. The result of clinical sample detection showed that the infection rate of FeChPV in 124 samples detected using qPCR assay was higher than that with conventional PCR. The established qPCR assay could be a low-cost, convenient, and reliable method to detect FeChPV in clinical practice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Feline parvovirus, also called feline panleukopenia virus (FPV), feline infectious enteritis virus, and feline plague virus, causes infectious diseases, such as high fever, vomiting, and serious leukopenia. It is characterized by a reduction in the number of circulating lymphocytes and enteritis (Tucciarone et al. 2021). Chaphamaparvovirus (ChPV) is a novel type of feline parvovirus.
ChPVs are a newly identified genus of parvoviruses (Penzes et al. 2020). Diarrhea in cats caused by an infectious agent has been reported since the 1930s. Diarrhea caused by infectious factors not only induces acute enterogastritis but also threatens the health of cats (Zhang et al. 2019). To date, feline astrovirus (FeAstV), feline bocavirus (FBoV), feline coronavirus (FCoV), feline kobuvirus (FKV), and feline parvovirus (FPV) can cause diarrhea in cats (Tasker 2018; Liu et al. 2018; Hoshino et al. 1981; Lu et al. 2018; Mietzsch et al. 2019).
Members of the Parvoviridae family are non-enveloped viruses with linear, single-stranded DNA genomes of 4–6 kb in length (Li et al. 2020; Penzes et al. 2019). According to the latest taxonomic criteria of the International Committee on Taxonomy of Viruses, the Parvoviridae family has been divided into three subfamilies: Densovirinae, Hamaparvovirinae, and Parvovirinae. The viruses in the Parvovirinae subfamily only infect vertebrates, and the Densovirinae subfamily uses invertebrates as hosts (Cotmore et al. 2014; Fahsbender et al. 2019). Hamaparvovirinae has a broad host range, including vertebrates and invertebrates. ChPV is one of the genera in Hamaparvovirinae and has been reported to infect various animals, such as dogs, cats, rats, barn owls, peafowls, mice, tilapias, red-crowned cranes, pigs, ducks, and chickens (Di Profio et al. 2021; Yang et al. 2016; Hargitai et al. 2021; Liu et al. 2020; Ge et al. 2020; Du et al. 2019; Wang et al. 2019, 2020a, b; Vibin et al. 2020; Lima et al. 2019; Mohamed et al, 2013). Feline Chaphamaparvovirus (FeChPV) was first identified in fecal samples of cats from animal shelters, where outbreaks of diarrhea and vomiting occurred in Canada in 2019 (Li et al. 2020). It has also been reported to be associated with diarrhea (Di Profio et al. 2021).
Few studies have been conducted on FeChPV, and currently, no efficient methods to detect FeChPV have been established. Therefore, a convenient method for FeChPV detection is urgently needed. The quantitative real-time polymerase chain reaction (qPCR) assay is deemed to be an accurate and effective method for virus detection (Liu et al. 2013; Chang et al. 2020). Therefore, in this study, a SYBR Green I-based qPCR assay was established for the clinical detection of FeChPV.
Materials and methods
Sample collection and nucleic acid extraction
Disease material source
In total, 20 cat fecal samples were collected from **n’an Animal Hospital in Hefei, Capital of East China’s Anhui Province, on October 25, 2019. Three of the samples were from cats with diarrhea, and 10 were from cats with normal medical examinations.
Treatment of disease material samples
All fecal samples were thoroughly ground in the grinder until no large particles were visible to the naked eye, and 1 mL phosphate-buffered saline buffer was added during the grinding process. All stool samples were evenly mixed and placed in a low-temperature centrifuge and centrifuged at 12,000 r/min for 10 min. Subsequently, the supernatant was taken from a sterile operating table and filtered with a 0.22 μm filter. The filtered liquid was placed in a 1.5 mL sterile centrifuge tube. All samples were stored at − 80℃ until use.
Extraction of viral nucleic acid
FeChPV DNA was extracted from fecal samples that were confirmed to be positive using the TIANamp Stool DNA kit (Tiangen, Bei**g, China). The strains of FeAstV (AH-1–2020 strain, GenBank number: MN977118), FBoV-1 (GenBank number: MT577646.1), FPV (HF1 strain, GenBank number: MT614366), FCoV (HF1902 strain, GenBank number: MT444152), feline calicivirus (FCV, ANHF1 strain, GenBank number: MT649084), and feline herpesvirus (FHV) (Fel-O-Vax PCT feline vaccine, Boehringer Ingelheim) were reserved in our laboratory.
Virus nucleic acid detection
FeChPV-specific primers were used for PCR detection of the samples. The reaction system used 20 μL conventional PCR system, using cDNA extracted from the pathogen as template, PCR amplification was performed with FeChPV-F and FeChPV-R primers at 95 ℃ for 5 min. 95 ℃ 30 s, 55 ℃ 30 s, 72 ℃ 40 s, 40 cycles; 72 ℃ for 10 min (for primer sequence, please refer to construction of standard plasmid). The negative control group was used for each test.
Construction of standard plasmid
To construct the standard plasmid, the DNA that was identified by sequencing as fechavirus was used as the template to amplify partial VP1 gene of 620 bp using a conventional PCR (cPCR) method. The forward (FeChPV-F) and reverse primers (FeChPV-R) were 5′-TGGGAACCAACAAACACG-3′ and 5′-CATCCTTGGGTAGTCGTT-3′, respectively. The DNA identified by sequencing as FeChPV was used as the template to amplify the partial VP1 gene. Furthermore, 10 μL 2 × Mix, 1 μL forward primer, 1 μL reverse primer, 2 μL template, and 6 μL double-distilled water (ddH2O) formed a system and were amplified using the cPCR method. After verification, the final conditions were set as follows: predegeneration at 95 °C for 5 min, followed by 40 cycles of degeneration at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 72 s, and a final extension step at 72 °C for 10 min. The PCR product was cloned into the pMD-19 T vector after purification and was named FeChPV-19 T. After transformation into DH5α cells, the Easy Pure Plasmid MiniPrep kit (TransGen Biotech, Bei**g, China) was used to extract the recombinant plasmid, and sequencing was performed by General Biosystems (Chuzhou, China).
After measurement using an ND-2000 spectrophotometer (Thermo Scientific, Dreieich, Germany) and calculation using the following formula: DNA concentration (copy number) = (6.02 × 1023 copies/mol × plasmid concentration [ng/μL] × 10−9)/(DNA length in nucleotides × 660 g/mol) were performed, the copy number of FeChPV DNA was determined. According to the copy number, the recombinant plasmid was tenfold diluted to 101.
Primer design
Based on the original research in our laboratory, we used the genome extracted by our laboratory to design primers through NCBI website, Primer5, Megalign and other software, carefully compared and repeatedly verified. Our reference sequence is FeChPV isolate HF2, complete genome (GenBank number: MT708231.1). A pair of specific primers was designed based on the regions that were used to construct the standard plasmid. The sequence of the forward primer was 5′-GCGTATACCGTATGGGGTCA-3′, and the reverse primer sequence was 5′-AGTCCCTGGGAATCTCCATC-3′. All primers used in this study were synthesized by General Biosystems.
Establishment of a standard curve
A tenfold serial dilution of recombinant plasmid was used as a template to perform a qPCR assay on a CFX96™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The system consisted of 10 μL of SuperReal PreMix Plus (Tiangen), 0.6 μL forward primer, 0.6 μL reverse primer, 1 μL template, and 7.8 μL ddH2O. The qPCR conditions were performed at 95 °C for 15 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s.
Sensitivity analysis
To evaluate the sensitivity of the qPCR assay, concentration gradients from 108 to 101 were used as templates. At the same time, cPCR was conducted using the same templates to compare the differences in sensitivity between the two methods.
Specificity analysis
To evaluate the specificity of qPCR, the DNA or cDNA of FPV, FeAstV, FBoV-1, FCV, FHV, and FCoV and a negative control of ddH2O were used as templates to observe the presence of specific curves.
Repeatability analysis
Four concentrations, 108, 106, 104, and 102, were selected to calculate the coefficients of Ct value variation (CV). The dilutions of the concentrations mentioned above were used as templates in the reaction system, each of which was performed three times a day and repeated for 3 days under the same conditions. The inter- and intra-assay CVs were then calculated to assess the reproducibility of the SYBR Green I-based assay.
Clinical sample detection
In total, 124 clinical samples collected from an animal hospital were used to verify whether the qRT-PCR assay was suitable for clinical practice. The DNA of the samples was extracted using the TIANamp Stool DNA kit (Tiangen) and stored at − 20 °C. Subsequently, qPCR and cPCR were used to detect the target viruses.
Results
Construction of the standard curve
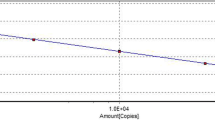
The cope number of FeChPV was 1.07 × 1011 copies/μL, and the dilutions used to establish the standard curve ranged from 1.07 × 1011 to 1.07 × 101 copies/μL. Ultimately, the standard curve that we received was y = − 3.330 × + 34.166, with a correlation coefficient (R2) of 0.999 and an amplification efficiency (E) of 99.7% (Fig. 1b). As shown in Fig. 1c, the melting curve showed a specific peak at 79 °C.
a Amplification curve of SYBR Green I-based quantitative real-time polymerase chain reaction (PCR). The copy numbers of feline Chaphamaparvovirus (FeChPV) DNA ranged from 1.07 × 108 to 1.07 × 101 copies/μL. b Standard curve of FeChPV (concentrations ranging from 1.07 × 108 to 1.07 × 101 copies/μL, y = − 3.330 × + 34.166, R2 = 0.999, E = 99.7%). c Melting curve of FeChPV (Tm = 79 ℃). d The gel electropherogram result of conventional PCR. DNA marker of 2000 bp was used. Lanes 1–8 underwent tenfold serial dilutions of the plasmids ranging from 1.07 × 108 to 1.07 × 101 copies/μL; Lane 9 was a negative control. The lowest limit of detection of conventional PCR was 1.07 × 103 copies/μL
Analysis of sensitivity for the qPCR
The result of sensitivity analysis showed that the limit concentration that the qPCR assay could detect was 1.07 × 101 (Fig. 2), whereas the cPCR was only able to detect up to 1.07 × 103 (Fig. 1d).
Analysis of specificity for the qPCR
As shown in Fig. 3, only FeChPV exhibited specific curves. The other viruses and ddH2O did not show specific curves.
Analysis of reproducibility for the qPCR
The inter- and intra-assay CVs were calculated to test the reproducibility of the qPCR assay. The inter-assay CVs ranged from 0.21 to 0.67%, and the intra-assay CVs ranged from 0.10 to 0.56% (Table 1).The formula for calculating the coefficient of variation is: coefficient of variation CV = (Standard Deviation SD/Mean Mean) × 100%.
Clinical samples testing by qPCR
As shown in Table 2, 124 stool samples were collected from animal hospitals and shelters in different areas of Anhui Province. After extracting nucleic acid from these materials, the qPCR method for FeChPV was established in this study for detection, and the same template was also detected by cPCR. The results showed that there were three positive samples detected by cPCR, with a positive rate of 2.42%, and six positive samples detected by qPCR, with a positive rate of 4.84%. The degree of agreement between the two methods was consistent, and all positive samples were verified by sequencing. This indicates that the sensitivity and specificity of the qPCR method established in this study can be applied to the clinical detection of FeChPV at the grassroots level.
Discussion
In clinical practice, FPV has been recognized as a general cause of feline diarrhea (Chang et al. 2020). Other viral pathogens related to enteritis are neglected to some extent (Zhang et al. 2019). Among NS1, VP1, and NP genes, the VP1 gene is more conserved. Therefore, based on the VP1 gene of FeChPV, we searched and analyzed a conserved region using Basic Local Alignment Search Tool (BLAST) in GenBank (GenBank numbers:MW404252.1, MW404253.1, MN794869.1). However, in the past few years, other viruses, including FCV (Di Martino et al. 2020), feline astrovirus (Wang et al. 2021a, b), and FBoV (Wang et al. 2021a, b), have been reported to be associated with diarrhea. In shelters where outbreaks of diarrhea and vomiting were observed, the detection rate of FeChPV was 47% in the fecal samples from 17 cats(Li et al. 2020), which indicated that FeChPV was a cause of diarrhea to a great degree.
A cPCR assay was performed when FeChPV was first found (Li et al. 2020). Previous studies on FeChPV have not provided any methods with the ability to rapidly detect the target virus. The TaqMan method has been described in articles related to enteritis and upper respiratory tract diseases (Wang et al. 2020a, b). The qPCR assay has a sensitivity and specificity that are greater than those obtained with the cPCR assay (Di Martino et al. 2020; Wang et al. 2020a, b, 2021a, b; Zheng et al. 2020). Given its low cost and convenient operation, the SYBR Green I assay is a better choice for large clinical sample testing.
At present, there is no official gold standard for FeChPV detection. According to the specific primers designed from the FeChPV genome, FeChPV can be accurately detected, but the sensitivity of traditional PCR methods is not very high. In this study, we sequenced all the samples that were initially positive, and finally determined that they were positive samples, but we cannot guarantee that there is no possibility of false negatives. In the present study, qPCR was successfully used to detect FeChPV in cats. The tenfold series dilutions from 1.07 × 108 to 1.07 × 101 copies/μL were used to obtain the standard curve of FeChPV. The standard curve equation was y = − 3.330 × + 34.166, with an R2 of 0.999 and an E of 99.7%. The sensitivity for FeChPV detection using the SYBR Green I assay was 100 times higher than that of cPCR. Thus, qPCR was confirmed to be accurate and sensitive. A few viruses related to diarrhea in cats and a negative control of ddH2O were used to assess whether the qPCR assay has reliable specificity, and the result suggested that it had good specificity. The reproducibility was also proven to be all-right with intra- and inter-assay CVs of 0.10–0.56% and 0.21–0.67%, respectively. To verify the practicability of the SYBR Green I assay in clinical practice, 124 fecal samples were tested for FeChPV positivity. In this study, we sequenced all the samples that were initially positive, and finally determined that they were positive samples, but we cannot guarantee that there is no possibility of false negatives. The results showed a higher detection rate for the target virus than for cPCR. This indicated that the qPCR assay is an efficient and accurate method for FeChPV detection. We found that one third of samples from animals clinically infected with FeChPV tested positive for FPV, FeAstV, and FBoV. We speculated that FeChPV may co-infect FPV, FeAstV, and FBoV. The potential impact of FeChPV on feline animals has been determined, and there is no effective prevention and treatment drug at present, which requires further study.
Conclusion
In summary, an SYBR Green I-based qPCR assay was successfully performed for FeChPV detection. The study has filled the gaps in the rapid detection of FeChPV and is appropriate for clinical diagnosis because it is sensitive, specific, and fast. This new detection method is of great significance to further study the epidemic trend of FeChPV in Anhui Province. Our findings contribute to further investigation of the prevalence, genotype distribution, and genetic diversity of FeChPV, adding to the molecular epidemiology of FeChPV worldwide.
References
Chang WS, Li CX, Hall J, Eden JS, Hyndman TH, Holmes EC et al (2020) Meta-transcriptomic discovery of a divergent circovirus and a chaphamaparvovirus in captive reptiles with proliferative respiratory syndrome. Viruses 12:108203
Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M et al (2014) The family Parvoviridae. Adv Virol 159:1239–1247
Di Martino B, Lanave G, Di Profio F, Melegari I, Marsilio F, Camero M et al (2020) Identification of feline calicivirus in cats with enteritis. Transbound Emerg Dis 67:2579–2588
Di Profio F, Sarchese V, Palombieri A, Fruci P, Massirio I, Martella V et al (2021) Feline chaphamaparvovirus in cats with enteritis and upper respiratory tract disease. Transbound Emerg Dis. https://doi.org/10.1111/tbed.14032
Du J, Wang W, Chan JF, Wang G, Huang Y, Yi Y et al (2019) Identification of a novel ichthyic parvovirus in marine species in Hainan Island, China. Front Microbiol 10:2815
Fahsbender E, Altan E, Seguin MA, Young P, Estrada M, Leutenegger C et al (2019) Chapparvovirus DNA found in 4% of dogs with diarrhea. Viruses 11:398
Ge Z, Carrasco SE, Feng Y, Bakthavatchalu V, Annamalai D, Kramer R et al (2020) Identification of a new strain of mouse kidney parvovirus associated with inclusion body nephropathy in immunocompromised laboratory mice. Emerg Microbes Infect 9:1814–1823
Hargitai R, Boros A, Pankovics P, Matics R, Altan E, Delwart E et al (2021) Detection and genetic characterization of a novel parvovirus (family Parvoviridae) in barn owls (Tyto alba) in Hungary. Adv Virol 166:231–236
Hoshino Y, Zimmer JF, Moise NS, Scott FW (1981) Detection of astroviruses in feces of a cat with diarrhea. Adv Virol 70:373–376
Li Y, Gordon E, Idle A, Altan E, Seguin MA, Estrada M et al (2020) Virome of a feline outbreak of diarrhea and vomiting includes bocaviruses and a novel chapparvovirus. Viruses 12:506
Lima DA, Cibulski SP, Tochetto C, Varela APM, Finkler F, Teixeira TF et al (2019) The intestinal virome of malabsorption syndrome-affected and unaffected broilers through shotgun metagenomics. Virus Res 261:9–20
Liu Z, Fu Y, Ji Y, Wei J, Cai X, Zhu Q (2013) Development and validation of one-step SYBR green real-time RT-PCR for the rapid detection of newly emerged duck Tembusu virus. Avian Dis 57:595–601
Liu C, Liu F, Li Z, Qu L, Liu D (2018) First report of feline bocavirus associated with severe enteritis of cat in Northeast China, 2015. J Vet Med Sci 80:731–735
Liu X, Wang H, Liu X, Li Y, Chen J, Zhang J et al (2020) Genomic and transcriptional analyses of novel parvoviruses identified from dead peafowl. Virology 539:80–91
Lu G, Zhang X, Luo J, Sun Y, Xu H, Huang J et al (2018) First report and genetic characterization of feline kobuvirus in diarrhoeic cats in China. Transbound Emerg Dis 65:1357–1363
Mietzsch M, Penzes JJ, Agbandje-McKenna M (2019) Twenty-five years of structural Parvovirology. Viruses 11:362
Mohamed N, Nilsson E, Johansson P, Klingstrom J, Evander M, Ahlm C et al (2013) Development and evaluation of a broad reacting SYBR-green based quantitative real-time PCR for the detection of different hantaviruses. J Clin Virol 56:280–285
Penzes JJ, de Souza WM, Agbandje-McKenna M, Gifford RJ (2019) An ancient lineage of highly divergent parvoviruses infects both vertebrate and invertebrate hosts. Viruses 11:525
Penzes JJ, Soderlund-Venermo M, Canuti M, Eis-Hubinger AM, Hughes J, Cotmore SF et al (2020) Reorganizing the family Parvoviridae: a revised taxonomy independent of the canonical approach based on host association. Adv Virol 165:2133–2146
Tasker S (2018) Diagnosis of feline infectious peritonitis: update on evidence supporting available tests. J Feline Med Surg 20:228–243
Tucciarone CM, Franzo G, Legnardi M, Lazzaro E, Zoia A, Petini M et al (2021) Genetic insights into feline parvovirus: evaluation of viral evolutionary patterns and association between phylogeny and clinical variables. Viruses 13:1033
Vibin J, Chamings A, Klaassen M, Bhatta TR, Alexandersen S (2020) Metagenomic characterisation of avian parvoviruses and picornaviruses from Australian wild ducks. Sci Rep 10:12800
Wang Y, Yang S, Liu D, Zhou C, Li W, Lin Y et al (2019) The fecal virome of red-crowned cranes. Adv Virol 164:3–16
Wang D, Mai J, Yang Y, Wang N (2020a) Porcine parvovirus 7: evolutionary dynamics and identification of epitopes toward vaccine design. Vaccines (basel) 8:359
Wang Y, Sun J, Guo X, Zhang D, Cui Y, Li W et al (2020b) TaqMan-based real-time polymerase chain reaction assay for specific detection of bocavirus-1 in domestic cats. Mol Cell Probes 53:101647
Wang Y, Li W, Guo X, Zhang D, Sun J, Fu Z et al (2021a) Development of SYBR Green I-based polymerase chain reaction for feline bocavirus 1 detection. 3 Biotech 11:61
Wang Y, Fu Z, Guo X, Zhang D, Bai C, Li W et al (2021b) Development of SYBR Green I-based real-time reverse transcription polymerase chain reaction for the detection of feline astrovirus. J Virol Methods 288:114012
Yang S, Liu Z, Wang Y, Li W, Fu X, Lin Y et al (2016) A novel rodent Chapparvovirus in feces of wild rats. Virol J 13:133
Zhang Q, Niu J, Yi S, Dong G, Yu D, Guo Y et al (2019) Development and application of a multiplex PCR method for the simultaneous detection and differentiation of feline panleukopenia virus, feline bocavirus, and feline astrovirus. Adv Virol 164:2761–2768
Zheng LL, Chai LY, Tian RB, Zhao Y, Chen HY, Wang ZY (2020) Simultaneous detection of porcine reproductive and respiratory syndrome virus and porcine circovirus 3 by SYBR Green capital I, Ukrainian-based duplex real-time PCR. Mol Cell Probes 49:101474
Acknowledgements
This research is funded by Anhui Agricultural University’s 2021 school-level undergraduate innovation and entrepreneurship project (project number: 202110364010). We would like to thank Editage (www.editage.cn) for English language editing.
Funding
Anhui Agricultural University’s 2021 school-level undergraduate innovation and entrepreneurship project (grant no. project number: 202110364010); Anhui Provincial College Students Innovation and entrepreneurship project (grant no. project number: s202110364018).
Author information
Authors and Affiliations
Contributions
XL: investigation, data curation, formal analysis, writing original draft. SL and XL: data curation, investigation. RW: data curation, investigation. XX and HW: funding acquisition, conceptualization, supervision, writing—review and editing.
Corresponding author
Ethics declarations
Conflict of interest
All authors have declared that there was no competing interests existing in the study.
Rights and permissions
About this article
Cite this article
Liu, X., Li, S., Liu, X. et al. Establishment of SYBR green I-based quantitative real-time polymerase chain reaction for the rapid detection of a novel Chaphamaparvovirus in cats. 3 Biotech 12, 91 (2022). https://doi.org/10.1007/s13205-022-03150-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03150-1