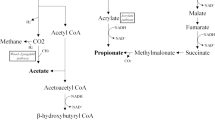
Abstract
Resistant starch (RS) is an important food source from which gut bacteria produce short chain fatty acids, which have beneficial effects for human health. The Bifidobacterium adolescentis P2P3, a human gut bacterium possessing a strong RS-degrading activity, was isolated from a healthy Korean adult male. In vitro experiments showed that this bacterium could utilize approximately 63% of high amylose corn starch after forming RS granule clusters. Here we provide the first complete set of genomic information on RS-degrading B. adolescentis P2P3. The genome of B. adolescentis P2P3 consists of one chromosome (2,202,982 bp) with high GC content (59.4%). Analysis of the protein-coding genes revealed that at least nineteen of the starch degradation-related enzymes were present in the genome. Among those, five genes evidently possess carbohydrate-binding domains, which are presumed to be involved in efficient RS decomposition. The complete set of genomic information on B. adolescentis P2P3 could provide an understanding of the role of RS-degrading gut bacteria and its RS degradation mechanism.



Similar content being viewed by others
References
Armenteros JJA, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420
Ashwar BA, Gani A, Shah A, Wani IA, Masoodi FA (2016) Preparation, health benefits and applications of resistant starch: a review. Starch-Starke 68:287–301
Bird A, Conlon M, Christophersen C, Top** D (2010) Resistant starch, large bowel fermentation and a broader perspective of prebiotics and probiotics. Benef Mirbobes 1:423–431
Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569
DuBois M, Gilles KA, Hamilton JK, Pt R, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356
Duranti S, Turroni F, Lugli GA, Milani C, Viappiani A, Mangifesta M, Gioiosa L, Palanza P, van Sinderen D, Ventura MJ (2014) Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22L. Appl Environ Microbiol 80:6080–6090
Grant JR, Stothard P (2008) The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184
Guillén D, Sánchez S, Rodríguez-Sanoja R (2010) Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biotechnol 85:1241–1249
Jung D-H, Seo D-H, Kim G-Y, Nam Y-D, Song E-J, Yoon S, Park C-S (2018) The effect of resistant starch (RS) on the bovine rumen microflora and isolation of RS-degrading bacteria. Appl Microbiol Biotechnol 102:4927–4936
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lee J, Ametani A, Enomoto A, Sato Y, Motoshima H, Ike F, Kaminogawa S (1993) Screening for the immunopotentiating activity of food microorganisms and enhancement of the immune response by Bifidobacterium adolescentis M101–4. Biosci Biotechnol Biochem 57:2127–2132
Lee I, Kim YO, Park S-C, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Litou ZI, Bagos PG, Tsirigos KD, Liakopoulos TD, Hamodrakas SJ (2008) Prediction of cell wall sorting signals in gram-positive bacteria with a hidden Markov model: application to complete genomes. J Bioinform Comput Biol 6:387–401
Liu S, Ren F, Zhao L, Jiang L, Hao Y, ** J, Zhang M, Guo H, Lei X, Sun E (2015) Starch and starch hydrolysates are favorable carbon sources for Bifidobacteria in the human gut. BMC Microbiol 15:54
Madeira F, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey A, Potter SC, Finn RD, Lopez R (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR (2016) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203
Masuko T, Minami A, Iwasaki N, Majima T, Nishimura S-I, Lee YC (2005) Carbohydrate analysis by a phenol–sulfuric acid method in microplate format. Anal Biochem 339:69–72
Morrison DJ, Preston T (2016) Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7:189–200
Mukhopadhya I, Moraïs S, Laverde-Gomez J, Sheridan PO, Walker AW, Kelly W, Klieve AV, Ouwerkerk D, Duncan SH, Louis P (2018) Sporulation capability and amylosome conservation among diverse human colonic and rumen isolates of the keystone starch-degrader Ruminococcus bromii. Environ Microbiol 20:324–336
Navarre WW, Schneewind O (1994) Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in Gram-positive bacteria. Mol Microbiol 14:115–121
Picard C, Fioramonti J, Francois A, Robinson T, Neant F, Matuchansky C (2005) Bifidobacteria as probiotic agents-physiological effects and clinical benefits. Aliment Pharmacol Ther 22:495–512
Rodriguez-Sanoja R, Ruiz B, Guyot J-P, Sanchez S (2005) Starch-binding domain affects catalysis in two Lactobacillus α-amylases. Appl Environ Microbiol 71:297–302
Ryan SM, Fitzgerald GF, van Sinderen D (2006) Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl Environ Microbiol 72:5289–5296
Saavedra JM, Bauman NA, Perman J, Yolken R, Oung I (1994) Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet 344:1046–1049
Southall SM, Simpson PJ, Gilbert HJ, Williamson G, Williamson MP (1999) The starch-binding domain from glucoamylase disrupts the structure of starch. FEBS Lett 447:58–60
Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278:631–637
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624
Ton-That H, Marraffini LA, Schneewind O (2004) Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta-Mol Cell Res 1694:269–278
Turroni F, Van Sinderen D, Ventura M (2011) Genomics and ecological overview of the genus Bifidobacterium. Int J Food Microbiol 149:37–44
Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F (2012) Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 7:e36957
Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D (2007) Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev 71:495–548
Ze X, Duncan SH, Louis P, Flint HJ (2012) Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6:1535–1543
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Acknowledgements
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (No. 2017R1A2B4004218).
Author information
Authors and Affiliations
Contributions
C-S P designed and coordinated all the experiments; D-H J performed the experiments, analyzed the genome data and wrote the manuscript; W-H C, Y-D N and D-H S performed the genome sequencing and sequence assembly; Y-J K measured residual RS. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of human derivatives were followed. The study of human derivatives was approved by the Institutional Review Board of Kyung Hee University (KHSIRB-17-004).
Additional information
Nucleotide sequence accession number: the complete chromosomal sequence of B. adolescentis P2P3 was deposited into Genbank under the Accession Number CP024959.
Rights and permissions
About this article
Cite this article
Jung, DH., Chung, WH., Seo, DH. et al. Complete genome sequence of Bifidobacterium adolescentis P2P3, a human gut bacterium possessing strong resistant starch-degrading activity. 3 Biotech 10, 31 (2020). https://doi.org/10.1007/s13205-019-2019-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-2019-7