Abstract
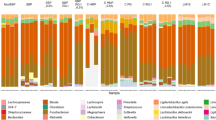
The gut microbiome plays a critical role to all animals and humans health. Methods based on ex vivo cultures are time and cost-effective solutions for rapid evaluation of probiotic effects on microbiomes. In this study, we assessed whether the protein secretome from the potential probiotic Enterococcus durans LAB18S grown on fructoligosaccharides (FOS) and galactoligosaccharides (GOS) had specific effects on ex vivo cultured intestinal microbiome obtained from a healthy individual. Metaproteomics was used to evaluate changes in microbial communities of the human intestinal microbiome. Hierarchical clustering analysis revealed 654 differentially abundant proteins from the metaproteome samples, showing that gut microbial protein expression varied on the presence of different E. durans secretomes. Increased amount of Bacteroidetes phylum was observed in treatments with secretomes from E. durans cultures on FOS, GOS and albumin, resulting in a decrease of the Firmicutes to Bacteroidetes (F/B) ratio. The most functionally abundant bacterial taxa were Roseburia, Bacteroides, Alistipes and Faecalibacterium. The results suggest that the secretome of E. durans may have favorable effects on the intestinal microbial composition, stimulating growth and different protein expression of beneficial bacteria. These findings suggest that proteins secreted by E. durans growing on FOS and GOS have different effects on the modulation of gut microbiota functional activities during cultivation.





Similar content being viewed by others
Availability of Data and Material
The proteomics mass spectrometry data are available via ProteomicXchange under the identifier PXD040751 (Username: reviewer_pxd040751@ebi.ac.uk; Password: glc8SRky).
References
Garcia-Mantrana I, Selma-Royo M, Alcantara C, Collado MC (2018) Shifts on gut microbiota associated to Mediterranean diet adherence and specific dietary intakes on general adult population. Front Microbiol 9:890
Fan Y, Pedersen O (2021) Gut microbiota in human metabolic health and disease. Nature Rev Microbiol 19:55–71
Cao Y, Oh J, Xue M, Huh WJ, Wang J, González-Hernández JA, Rice TA, Martin AL, Song D, Crawford JM, Herzon SB, Palm NW (2022) Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science 378:eabm3233
Galloway-Peña J, Hanson B (2020) Tools for analysis of the microbiome. Digest Dis Sci 65:674–685
Mayne J, Ning Z, Zhang X, Starr AE, Chen R, Deeke S, Chiang CK, Xu B, Wen M, Cheng K, Seebun D, Star A, Moore JI, Figeys D (2016) Bottom-up proteomics (2013–2015): kee** up in the era of systems biology. Anal Chem 88:95–121
Stamboulian M, Canderan J, Ye Y (2022) Metaproteomics as a tool for studying the protein landscape of human-gut bacterial species. PLoS Comput Biol 18:e1009397
Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME (2014) Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nature Rev Gastroenterol Hepatol 11:506–514
Clarke TC, Black LI, Stussman BJ, Barnes PM, Nahin RL (2015) Trends in the use of complementary health approaches among adults: United States, 2002–2012. Natl Health State Rep 79:1–16
Draper K, Ley C, Parsonnet J (2017) Probiotic guidelines and physician practice: a cross-sectional survey and overview of the literature. Beneficial Microbes 8:507–519
Irwin C, Khalesi S, Cox AJ, Grant G, Davey AK, Bulmer AC, Desbrow B (2017) Effect of 8-weeks prebiotics/probiotics supplementation on alcohol metabolism and blood biomarkers of healthy adults: a pilot study. Eur J Nutr 57:1523–1534
Kristensen NB, Bryrup T, Allin KH, Nielsen T, Hansen TH, Pedersen O (2016) Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med 8:52
Pieniz S, Andreazza R, Anghinoni T, Camargo FAO, Brandelli A (2014) Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18S. Food Control 37:251–256
Comerlato CB, Prichula J, Siqueira FM, Ritter AC, Varela APM, Mayer FQ, Brandelli A (2022) Genomic analysis of Enterococcus durans LAB18S, a potential probiotic strain isolated from cheese. Genet Mol Biol 45:e20210201
Comerlato CB, Zhang X, Walker K, Brandelli A, Figeys D (2020) Comparative proteomic analysis reveals metabolic variability of probiotic Enterococcus durans during aerobic and anaerobic cultivation. J Proteomics 220:103764
Li L, Ning Z, Zhang X, Mayne J, Cheng K, Stintzi A, Figeys D (2020) RapidAIM: a culture-and metaproteomics-based Rapid Assay of Individual Microbiome responses to drugs. Microbiome 8:33
Rossi M, Corradini C, Amaretti A, Nicolini M, Pompei A, Zanoni S, Matteuzzi D (2005) Fermentation of fructooligosaccharides and inulin by Bifidobacteria: a comparative study of pure and fecal cultures. Appl Environ Microbiol 71:6150–6158
Deeke SA, Starr AE, Ning Z, Ahamadi S, Zhang X, Mayne J, Chiang CK, Singleton R, Benchimol EI, Mack DR, Stintzi A, Figeys D (2018) Mucosal-luminal interface proteomics reveals biomarkers of pediatric inflammatory bowel disease-associated colitis. Am J Gastroenterol 113:713–724
Zhang X, Ning Z, Mayne J, Moore JI, Li J, Butcher J, Deeke SA, Chen R, Chiang CK, Wen M, Mack D, Stintzi A, Figeys D (2016) MetaPro-IQ: a universal metaproteomic approach to studying human and mouse gut microbiota. Microbiome 4:31
Cheng K, Ning Z, Zhang X, Li L, Liao B, Mayne J, Stintzi A, Figeys D (2017) MetaLab: an automated pipeline for metaproteomic data analysis. Microbiome 5:157
Singh RG, Tanca A, Palomba A, Van der Jeugt F, Verschaffelt P, Uzzau S, Martens L, Dawyndt P, Mesuere B (2019) Unipept 4.0: Functional analysis of metaproteome data. J Proteome Res 18:606–615
Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, **a J (2018) MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46:486–494
Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230
Lye HS, Kato T, Low WY (2017) Lactobacillus fermentum FTDC 8312 combats hypercholesterolemia via alteration of gut microbiota. J Biotechnol 262:75–83
Nakamoto N, Amiya T, Aoki R (2017) Commensal Lactobacillus controls immune tolerance during acute liver injury in mice. Cell Rep 21:1215–1226
Schmidt TSB, Raes J, Bork P (2018) The human gut microbiome: from association to modulation. Cell 172:1198–1215
Stojanov S, Berlec A, Štrukelj B (2020) The influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 8:1715
Magne F, Gotteland M, Gauthier L, Zazueta A, Pesoa S, Navarrete P, Balamurugan R (2020) The Firmicutes/Bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients 12:1474
Pieniz S, Moura TM, Cassenego AP, Andreazza R, Frazzon APG, Camargo FAO, Brandelli A (2015) Evaluation of resistance genes and virulence factors in a food isolated Enterococcus durans with probiotic potential. Food Control 51:49–54
Kitazawa H, Alvarez S, Suvorov A, Melnikov V, Villena J, Sánchez B (2015) Recent advances and future perspective in microbiota and probiotics. BioMed Res Int 2015:275631
Yu L, Zhao XK, Cheng ML (2017) Saccharomyces boulardii administration changes gut microbiota and attenuates D-galactosamine-induced liver injury. Sci Rep 7:1359
Chua MC, Ben-Amor K, Lay C (2017) Effect of synbiotic on the gut microbiota of cesarean delivered infants: a randomized, double-blind, multicenter study. J Pediatr Gastroenterol Nutr 65:102–106
Kosuwon P, Lao-Araya M, Uthaisangsook S (2018) A synbiotic mixture of scGOS/lcFOS and Bifidobacterium brevis M-16V increases faecal Bifidobacterium in healthy young children. Beneficial Microbes 9:541–552
De Filippis F, Esposito A, Ercolini D (2022) Outlook on next-generation probiotics from the gut. Cell Mol Life Sci 79:76
Deleu S, Machiels K, Raes J, Verbeke K, Vermeire S (2021) Short chain fatty acids and its producing organisms: an overlooked therapy for IBD? EBioMedicine 66:103293
Yan J, Pan Y, Shao W, Wang C, Wang R, He Y, Zhang M, Wang Y, Li T, Wang Z, Liu W, Wang Z, Sun X, Dong S (2022) Beneficial effect of the short-chain fatty acid propionate on vascular calcification through intestinal microbiota remodeling. Microbiome 10:195
He X, Zhao S, Li Y (2021) Faecalibacterium prausnitzii: a next-generation probiotic in gut disease improvement. Can J Infect Dis Med Microbiol 2021:6666114
Maioli TU, Borras-Nogues E, Torres L, Barbosa SC, Martins VD, Langella P, Azevedo VA, Chatel JM (2021) Possible benefits of Faecalibacterium prausnitzii for obesity-associated gut disorders. Front Pharmacol 12:740636
Kuda T, Brandelli A (2022) Effect of food ingredients on susceptible gut indigenous bacteria. In: Brandelli A (ed) Probiotics - advanced food and health applications. Academic Press, London 167–184
Sun M, Wu W, Liu Z, Cong Y (2017) Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol 52:1–8
LeBlanc JG, Chain F, Martín R, Bermúdez-Humarán LG, Courau S, Langella P (2017) Beneficial effects on host energy metabolism of short chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Factories 16:79
Yoshi K, Hosomi K, Kunisawa J (2019) Metabolism of dietary and microbial vitamin B family in the regulation of host immunity. Front Nutr 6:48
Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M (2020) The controversial role of human gut Lachnospiraceae. Microorganisms 8:573
Oliphant K, Ali M, D’Souza M, Hughes PD, Sulakhe D, Wang AZ, **e B, Yeasin R, Msall ME, Andrews B, Claud EC (2021) Bacteroidota and Lachnospiraceae integration into the gut microbiome at key time points in early life are linked to infant neurodevelopment. Gut Microbes 13:e1997560
Benus RF, van der Werf TS, Welling GW, Judd PA, Taylor MA, Harmsen HJ, Whelan K (2010) Association between Faecalibacterium prausnitzii and dietary fibre in colonic fermentation in healthy human subjects. Br J Nutr 104:693–700
Zafar H, Saier MH Jr (2021) Gut Bacteroides species in health and disease. Gut Microbes 13:1
Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A (2020) The genus Alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol 11:906
Rau M, Rehman A, Dittrich M, Groen AK, Hermanns HM, Seyfried F, Beyersdorf N, Rosenstiel P, Geier A (2018) Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United Eur Gastroenterol J 6:1496–1507
Medawar E, Haange SB, Rolle-Kampczyk U, Engelmann B, Dietrich A, Thieleking R, Wiegank C, Fries C, Horstmann A, Villringer A, von Bergen M, Fenske W, Witt AV (2021) Gut microbiota link dietary fiber intake and short-chain fatty acid metabolism with eating behavior. Transl Psychiatry 11:500
Gomez-Arango LF, Barrett HL, Wilkinson SA, Callaway LK, McIntyre HD, Morrison M, Nitert MD (2018) Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes 9:189–201
Astbury S, Atallah E, Vijay A, Aithal GP, Grove JI, Valdes AM (2020) Lower gut microbiome diversity and higher abundance of proinflammatory genus Collinsella are associated with biopsy-proven nonalcoholic steatohepatitis. Gut Microbes 11:569–580
Hirayama M, Nishiwaki H, Hamaguchi T, Ito M, Ueyama J, Maeda T, Kashihara K, Tsuboi Y, Ohno K (2021) Intestinal Collinsella may mitigate infection and exacerbation of COVID-19 by producing ursodeoxycholate. PLoS ONE 16:e0260451
Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI (2008) Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213–223
Ballan R, Battistini C, Xavier-Santos D, Saad SMI (2020) Interaction of probiotics and prebiotics with the gut microbiota. Progr Mol Biol Translat Sci 171:265–300
Watanabe Y, Nagai F, Morotomi M (2012) Characterization of Phascolarctobacterium succinatutens sp. nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl Environ Microbiol 78:511–518
Fernández-Veledo S, Vendrell J (2019) Gut microbiota-derived succinate: friend or foe in human metabolic diseases? Rev Endocr Metab Disord 20:439–447
Vidal-Veuthey B, González D, Cárdenas JP (2022) Role of microbial secreted proteins in gut microbiota-host interactions. Font Cell Infect Microbiol 12:964710
Zhang T, Zhang W, Feng C, Kwok LY, He Q, Sun Z (2022) Stronger gut microbiome modulatory effects by postbiotics than probiotics in a mouse colitis model. Npj Sci Food 6:53
Funding
This work received financial support from Conselho Nacional de Desenvolvimento Científico e Tecnológico of Brazil (CNPq, grant 308880/2021–8) and the Emerging Leaders in the Americas Program (ELAP, Canada). This work was also supported by the Government of Canada through the Natural Sciences and Engineering Research Council of Canada (NSERC, grant no. 210034). CBC was a former recipient of a PhD fellowship from CAPES.
Author information
Authors and Affiliations
Contributions
CBC, XZ, JM, AB and DF contributed to the study conception and design. Material preparation, data collection, CBC and KW; data analysis and statistical analysis were performed by CBC, XZ, JM. The manuscript was written by CBC, and critically revised by AB. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
The protocol for stool sample collection was approved by the Ottawa Health Science Network Research Ethics Board at the Ottawa Hospital (Ottawa, Canada).
Competing Interests
DF is a co-founder of MedBiome Inc., a personalized microbiome therapeutic company. The other Authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Comerlato, C.B., Zhang, X., Walker, K. et al. The Influence of Protein Secretomes of Enterococcus durans on ex vivo Human Gut Microbiome. Probiotics & Antimicro. Prot. (2023). https://doi.org/10.1007/s12602-023-10136-9
Accepted:
Published:
DOI: https://doi.org/10.1007/s12602-023-10136-9