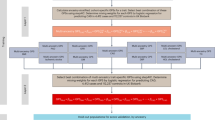
Abstract
We evaluated the performance of various polygenic risk score (PRS) models derived from European (EU), South Asian (SA), and Punjabi Asian Indians (AI) studies on 13,974 subjects from AI ancestry. While all models successfully predicted Coronary artery disease (CAD) risk, the AI, SA, and EU + AI were superior predictors and more transportable than the EU model; the predictive performance in training and test sets was 18% and 22% higher in AI and EU + AI models, respectively than in EU. Comparing individuals with extreme PRS quartiles, the AI and EU + AI captured individuals with high CAD risk showed 2.6 to 4.6 times higher efficiency than the EU. Interestingly, including the clinical risk score did not significantly change the performance of any genetic model. The enrichment of diversity variants in EU PRS improves risk prediction and transportability. Establishing population-specific normative and risk factors and inclusion into genetic models would refine the risk stratification and improve the clinical utility of CAD PRS.
Graphical abstract



Similar content being viewed by others
Data Availability
A part of the AIDHS/SDS data used in this study has already been deposited on the DbGap repository of the National Institutes of Health (https://www.ncbi.nlm.nih.gov/gap/). The additional datasets used and/or analyzed during the current study would be available from the corresponding author on reasonable request.
References
Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, Barengo NC, Beaton AZ, Benjamin EJ, Benziger CP, et al. Global burden of cardiovascular diseases and risk factors, 1990–2019: update from the GBD 2019 study. J Am Coll Cardiol. 2020;76(25):2982–3021.
Joshi P, Islam S, Pais P, Reddy S, Dorairaj P, Kazmi K, Pandey MR, Haque S, Mendis S, Rangarajan S, et al. Risk factors for early myocardial infarction in South Asians compared with individuals in other countries. JAMA : J Am Med Assoc. 2007;297(3):286–94.
Volgman AS, Palaniappan LS, Aggarwal NT, Gupta M, Khandelwal A, Krishnan AV, Lichtman JH, Mehta LS, Patel HN, Shah KS, et al. Atherosclerotic cardiovascular disease in South Asians in the United States: Epidemiology, risk factors, and treatments: A scientific statement from the American heart association. Circulation. 2018;138(1):e1–34.
Wang M, Menon R, Mishra S, Patel AP, Chaffin M, Tanneeru D, Deshmukh M, Mathew O, Apte S, Devanboo CS, et al. Validation of a genome-wide polygenic score for coronary artery disease in South Asians. J Am Coll Cardiol. 2020;76(6):703–14.
Gertler MM, Garn SM, White PD. Young candidates for coronary heart disease. J Am Med Assoc. 1951;147(7):621–5.
Benn M, Watts GF, Tybjaerg-Hansen A, Nordestgaard BG. Mutations causative of familial hypercholesterolaemia: screening of 98 098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur Heart J. 2016;37(17):1384–94.
Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330(15):1041–6.
Musunuru K, Kathiresan S. Genetics of common, complex coronary artery disease. Cell. 2019;177(1):132–45.
Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–24.
Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, Lai FY, Kaptoge S, Brozynska M, Wang T, et al. Genomic risk prediction of coronary artery disease in 480,000 Adults: Implications for primary prevention. J Am Coll Cardiol. 2018;72(16):1883–93.
Chatterjee N, Shi J, Garcia-Closas M. Develo** and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17(7):392–406.
Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584–91.
Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177(4):1080.
Moorjani P, Thangaraj K, Patterson N, Lipson M, Loh PR, Govindaraj P, Berger B, Reich D, Singh L. Genetic evidence for recent population mixture in India. Am J Hum Genet. 2013;93(3):422–38.
Sanghera DK, Been LF, Ralhan S, Wander GS, Mehra NK, Singh JR, Ferrell RE, Kamboh MI, Aston CE. Genome-wide linkage scan to identify loci associated with type 2 diabetes and blood lipid phenotypes in the Sikh Diabetes Study. PLoS One. 2011;6(6):e21188.
Saxena R, Saleheen D, Been LF, Garavito ML, Braun T, Bjonnes A, Young R, Ho WK, Rasheed A, Frossard P, et al. Genome-wide association study identifies a novel locus contributing to type 2 diabetes susceptibility in Sikhs of Punjabi origin from India. Diabetes. 2013;62(5):1746–55.
Sapkota BR, Hopkins R, Bjonnes A, Ralhan S, Wander GS, Mehra NK, Singh JR, Blackett PR, Saxena R, Sanghera DK. Genome-wide association study of 25(OH) Vitamin D concentrations in Punjabi Sikhs: Results of the Asian Indian diabetic heart study. J Steroid Biochem Mol Biol. 2016;158:149–56.
Sanghera DK, Bhatti JS, Bhatti GK, Ralhan SK, Wander GS, Singh JR, Bunker CH, Weeks DE, Kamboh MI, Ferrell RE. The Khatri Sikh Diabetes Study (SDS): Study design, methodology, sample collection, and initial results. Hum Biol. 2006;78(1):43–63.
Sanghera DK, Hopkins R, Malone-Perez MW, Bejar C, Tan C, Mussa H, Whitby P, Fowler B, Rao CV, Fung KA, et al. Targeted sequencing of candidate genes of dyslipidemia in Punjabi Sikhs: Population-specific rare variants in GCKR promote ectopic fat deposition. PLoS One. 2019;14(8):e0211661.
Goyal S, Tanigawa Y, Zhang W, Chai JF, Almeida M, Sim X, Lerner M, Chainakul J, Ramiu JG, Seraphin C, et al. APOC3 genetic variation, serum triglycerides, and risk of coronary artery disease in Asian Indians, Europeans, and other ethnic groups. Lipids Health Dis. 2021;20(1):113.
Rout M, Lerner M, Blackett PR, Peyton MD, Stavrakis S, Sidorov E, Sanghera DK. Ethnic differences in ApoC-III concentration and the risk of cardiovascular disease: No evidence for the cardioprotective role of rare/loss of function APOC3 variants in non-Europeans. Am Heart J Plus. 2022;13:100128.
Braun TR, Been LF, Blackett PR, Sanghera DK. Vitamin D Deficiency and cardio-metabolic risk in a North Indian community with highly prevalent type 2 diabetes. J Diabetes Metab. 2012;3:2155–6156.
American Diabetes A. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2013;36(Suppl 1):S67-74.
Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, Mehra NK, Mulvihill JJ, Ferrell RE, Nath SK, et al. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet. 2008;9:59.
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779.
Sun L, Pennells L, Kaptoge S, Nelson CP, Ritchie SC, Abraham G, Arnold M, Bell S, Bolton T, Burgess S, et al. Polygenic risk scores in cardiovascular risk prediction: A cohort study and modelling analyses. PLoS Med. 2021;18(1):e1003498.
Manikpurage HD, Eslami A, Perrot N, Li Z, Couture C, Mathieu P, Bosse Y, Arsenault BJ, Theriault S. Polygenic risk score for coronary artery disease improves the prediction of early-onset myocardial infarction and mortality in men. Circ Genom Precis Med. 2021;14(6):e003452.
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215.
Evangelou E, Ioannidis JP. Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet. 2013;14(6):379–89.
Saxena R, Bjonnes A, Prescott J, Dib P, Natt P, Lane J, Lerner M, Cooper JA, Ye Y, Li KW, et al. Genome-wide association study identifies variants in casein kinase II (CSNK2A2) to be associated with leukocyte telomere length in a Punjabi Sikh diabetic cohort. Circ Cardiovasc Genet. 2014;7(3):287–95.
Myocardial Infarction G, Investigators CAEC, Stitziel NO, Stirrups KE, Masca NG, Erdmann J, Ferrario PG, Konig IR, Weeke PE, Webb TR, et al. Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N Engl J Med. 2016;374(12):1134–44.
Webb TR, Erdmann J, Stirrups KE, Stitziel NO, Masca NG, Jansen H, Kanoni S, Nelson CP, Ferrario PG, Konig IR, et al. Systematic evaluation of pleiotropy identifies 6 further loci associated with coronary artery disease. J Am Coll Cardiol. 2017;69(7):823–36.
Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang W, Frossard P, Been LF, Chia KS, Dimas AS, Hassanali N, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43(10):984–9.
Zhao W, Rasheed A, Tikkanen E, Lee JJ, Butterworth AS, Howson JMM, Assimes TL, Chowdhury R, Orho-Melander M, Damrauer S, et al. Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat Genet. 2017;49(10):1450–7.
Mahajan A, Go MJ, Zhang WH, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MCY, Prokopenko I, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nature Genet. 2014;46(3):234.
Coronary Artery Disease Genetics C. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43(4):339–44.
Choi SW, Mak TS, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020;15(9):2759–72.
Lu X, Liu Z, Cui Q, Liu F, Li J, Niu X, Shen C, Hu D, Huang K, Chen J, et al. A polygenic risk score improves risk stratification of coronary artery disease: a large-scale prospective Chinese cohort study. Eur Heart J. 2022;43(18):1702–11.
Kannel WB, McGee D, Gordon T. A general cardiovascular risk profile: the Framingham Study. Am J Cardiol. 1976;38(1):46–51.
D’Agostino RB Sr, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, Kannel WB. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation. 2008;117(6):743–53.
Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1.
Singh U. A history of ancient and early medieval India : from the stone age to the 12th century. New Delhi ; Upper Saddle River, NJ: Pearson Education 2009.
Mascarenhas DD, Raina A, Aston CE, Sanghera DK. Genetic and Cultural Reconstruction of the Migration of an Ancient Lineage. Biomed Res Int. 2015;2015:651415.
Indian Genome Variation C. Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet. 2008;87(1):3–20.
Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461(7263):489–94.
Bejar CA, Goyal S, Afzal S, Mangino M, Zhou A, van der Most PJ, Bao Y, Gupta V, Smart MC, Walia GK, et al. A bidirectional mendelian randomization study to evaluate the causal role of reduced blood vitamin D levels with type 2 diabetes risk in South Asians and Europeans. Nutr J. 2021;20(1):71.
Venkatesan V, Lopez-Alvarenga JC, Arya R, Ramu D, Koshy T, Ravichandran U, Ponnala AR, Sharma SK, Lodha S, Sharma KK et al. Burden of type 2 diabetes and associated Cardiometabolic traits and their heritability estimates in endogamous ethnic groups of India: Findings from the Indigenius Consortium. Front Endocrinol 2022;13
Dron JS, Wang J, Cao HN, McIntyre AD, Iacocca MA, Menard JR, Movsesyan I, Malloy MJ, Pullinger CR, Kane JP, et al. Severe hypertriglyceridemia is primarily polygenic. J Clin Lipidol. 2019;13(1):80–8.
Hense HW, Schulte H, Lowel H, Assmann G, Keil U. Framingham risk function overestimates risk of coronary heart disease in men and women from Germany–results from the MONICA Augsburg and the PROCAM cohorts. Eur Heart J. 2003;24(10):937–45.
Dikilitas O, Schaid DJ, Tcheandjieu C, Clarke SL, Assimes TL, Kullo IJ. Use of polygenic risk scores for coronary heart disease in ancestrally diverse populations. Curr Cardiol Rep. 2022;24(9):1169–77.
Agbaedeng TA, Noubiap JJ, Mofo Mato EP, Chew DP, Figtree GA, Said MA, van der Harst P. Polygenic risk score and coronary artery disease: A meta-analysis of 979,286 participant data. Atherosclerosis. 2021;333:48–55.
Palmer C, Pe’er I. Statistical correction of the Winner’s Curse explains replication variability in quantitative trait genome-wide association studies. PLoS Genet. 2017;13(7):e1006916.
Acknowledgements
The Asian Indian Diabetic Heart Study/Sikh Diabetes Study was supported by National Institute of Health grants-R01DK082766 and R01DK118427 (National Institute of Diabetes and Digestive and Kidney Diseases, NIDDK) and the Presbyterian Health Foundation grants. The authors thank all the participants of AIDHS/SDS and are grateful for their contribution to this study. Technical support and genotype data generation by Adam Adler from the Oklahoma Medical Research Foundation are duly acknowledged.
Funding
The Asian Indian Diabetic Heart Study/Sikh Diabetes Study was supported by National Institute of Health grants-R01DK082766 and R01DK118427 (National Institute of Diabetes and Digestive and Kidney Diseases, NIDDK) and the Presbyterian Health Foundation grants.
Author information
Authors and Affiliations
Contributions
MR and GKT performed data analysis and helped in manuscript preparation; JRS, NKM, GSW, and SR contributed to recruitment and phenoty** for AIDHS/SDS. DKS designed the study, contributed to genoty** and phenoty** as a cohort PI of AIDHS/SDS, and wrote the manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
The study was approved by the University of Oklahoma Health Sciences Center Institutional Review Board (IRB#2911), as well as the Human Subject Protection Committee at the participating hospitals and institutes in India and informed consent was obtained from all the participants included in the study. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000 (5).
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Associate Editor Paul J. R. Barton oversaw the review of this article
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rout, M., Tung, G.K., Singh, J.R. et al. Polygenic Risk Score Assessment for Coronary Artery Disease in Asian Indians. J. of Cardiovasc. Trans. Res. (2024). https://doi.org/10.1007/s12265-024-10511-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12265-024-10511-z