Abstract
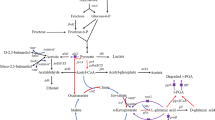
A breeding approach combining genome shuffling with streptomycin resistance was developed in this research to improve the poly-γ-L-diaminobutanoic acid (γ-PAB) production in Bacillus pumilus LS-1. By this unique strategy, recombinants from the third round of genome shuffling could tolerate 18 µg/L of streptomycin and exhibited higher γ-PAB yield as 152.2 mg/L in shake-flask fermentation, 3-fold over the parent. In batch fermentation, B. pumilus GS3-9 could produce γ-PAB as high as 1284.7 mg/L in two days, 2.4-fold higher than the control, which was the highest productivity ever reported. In addition, the optimal pH in B. pumilus for γ-PAB synthesis was changed after atmospheric and room temperature plasma (ARTP) mutagenesis and protoplast fusion, because lower pH environment is favorable for accumulation of intracellular ATP. Some key enzymes in GS3-9 showed higher activities than those in parent, suggesting a greater flux to TCA circle and DAP pathway, which was a reason for enhanced γ-PAB production.
Similar content being viewed by others
References
Kunioka, M. (1997) Biosynthesis and chemical reactions of poly(amino acid)s from microorganisms. Appl. Microbiol. Biotechnol. 47: 469–475.
Yamanaka, K., C. Maruyama, H. Takagi, and Y. Hamano (2008) Epsilon-poly-l-lysine dispersity is controlled by a highly unusual nonribosomal peptide synthetase. Nat. Chem. Biol. 4: 766–772.
Obst, M. and A. Steinbüchel (2004) Microbial degradation of poly(amino acid)s. Biomacromolecules. 5: 1166–1176.
Hamano, Y., T. Arai, M. Ashiuchi, and K. Kino (2013) NRPSs and amide ligases producing homopoly(amino acid)s and homooligo(amino acid)s. Nat. Prod. Rep. 30: 1087–1097.
Hiraki, J., T. Ichikawa, S. Ninomiya, H. Seki, K. Uohama, H. Seki, S. Kimura, Y. Yanagimoto, and J. W. Barnett Jr (2003) Use of ADME studies to confirm the safety of epsilon-polylysine as a preservative in food. Regul. Toxicol. Pharm. 37: 328–340.
**a, J., H. Xu, X. Feng, Z. Xu, and B. Chi (2013) Poly(L-diaminopropionic acid), a novel non-proteinic amino acid oligomer co-produced with poly(ε-l-lysine) by Streptomyces albulus PD-1. Appl. Microbiol. Biotechnol. 97: 7597–7605.
Takehara, M., M. Saimura, H. Inaba, and H. Hirohara (2008) Poly(γ-L-diaminobutanoic acid), a novel poly(amino acid), coproduced with poly(ε-L-lysine) by two strains of Streptomyces celluloflavus. FEMS. Microbiol. Lett. 286: 110–117.
Xu, Z., Z. Sun, S. Li, Z. Xu, C. Cao, Z. Xu, X. Feng, and H. Xu (2015) Systematic unravelling of the biosynthesis of poly(L-diaminopropionic acid) in Streptomyces albulus PD-1. Sci. Rep. 5: 17400.
Zhang, Y. X., K. Perry, V. A. Vinci, K. Powell, W. P. C. Stemmer, and S. B. del Cardayre (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 415: 644–646.
Kim, J. Y., H. W. Yoo, P. G. Lee, S. G. Lee, J. H. Seo, and B. G. Kim (2019) In vivo protein evolution, next generation protein engineering strategy: from random approach to target-specific approach. Biotechnol. Bioprocess Eng. 24: 85–94.
Patnaik, R., S. Louie, V. Gavrilovic, K. Perry, W. P. C. Stemmer, C. M. Ryan, and S. del Cardayré (2002) Genome shuffling of Lactobacillus for improved acid tolerance. Nat. Biotechnol. 20: 707–712.
Xu, B., Z. H. **, H. Wang, Q. **, X. **, and P. Cen (2008) Evolution of Streptomyces pristinaespiralis for resistance and production of pristinamycin by genome shuffling. Appl. Microbiol. Biotechnol. 80: 261–267.
Shi, D. J., C. L. Wang, and K. M. Wang (2009) Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 36: 139–147.
Zheng, D. Q., X. C. Wu, P. M. Wang, X. Q. Chi, X. L. Tao, P. Li, X. H. Jiang, and Y. H. Zhao (2011) Drug resistance marker-aided genome shuffling to improve acetic acid tolerance in Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 38: 415–422.
Li, S., F. Li, X. S. Chen, L. Wang, J. Xu, L. Tang, and Z. G. Mao (2012) Genome shuffling enhanced e-poly-L-lysine production by improving glucose tolerance of Streptomyces graminearus. Appl. Biochem. Biotechnol. 166: 414–423.
Wang, L., X. Chen, G. Wu, X. Zeng, X. Ren, S. Li, L. Tang, and Z. Mao (2016) Genome shuffling and gentamicin-resistance to improve e-poly-L-lysine productivity of Streptomyces albulus W-156. Appl. Biochem. Biotechnol. 180: 1601–1617.
Hu, H. and K. Ochi (2001) Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl. Environ. Microbiol. 67: 1885–1892.
Ochi, K., S. Okamoto, Y. Tozawa, T. Inaoka, T. Hosaka, J. Xu, and K. Kurosawa (2004) Ribosome engineering and secondary metabolite production. Adv. Appl. Microbiol. 56: 155–184.
Wang, L., S. Li, J. Zhao, Y. Liu, X. Chen, L. Tang, and Z. Mao (2019) Efficiently activated ε-poly-L-lysine production by multiple antibiotic-resistance mutations and acidic pH shock optimization in Streptomyces albulus. Microbiologyopen. 8: e00728.
Banerjee, S., G. Mishra, and A. Roy (2019) Metabolic engineering of bacteria for renewable bioethanol production from cellulosic biomass. Biotechnol. Bioprocess Eng. 24: 713–733.
Zeng, X., X. S. Chen, X. D. Ren, Q. R. Liu, L. Wang, Q. X. Sun, L. Tang, and Z. G. Mao (2014) Insights into the role of glucose and glycerol as a mixed carbon source in the improvement of epsilon-poly-L-lysine productivity. Appl. Biochem. Biotechnol. 173: 2211–2224.
Teichgraber, P., D. Biesold, and Z. D. Pigareva (1973) Subcellular localization of hexokinase in the rat cortex. Neurosci. Behav. Physiol. 6: 218–227.
Morris, C. N., S. Ainsworth, and J. Kinderlerer (1986) The regulatory properties of yeast pyruvate kinase. Effect of fructose 1,6-bisphosphate. Biochem. J. 234: 691–698.
Bradford, M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254.
Oda, Y., S. Nakamura, I. Oki, T. Kato, and H. Shinagawa (1985) Evaluation of the new system (umu-test) for the detection of environmental mutagens and carcinogens. Mutat. Res. 147: 219–229.
Wang, L. Y., Z. L. Huang, G. Li, H. X. Zhao X. H. **ng, W. T. Sun, H. P. Li, Z. X. Gou, and C. Y. Bao (2010) Novel mutation breeding method for Streptomyces avermitilis using an atmospheric pressure glow discharge plasma. J. Appl. Microbiol. 108: 851–858.
Xu, F., J. Wang, S. Chen, W. Qin, Z. Yu, H. Zhao, X. **ng, and H. Li (2011) Strain improvement for enhanced production of cellulase in Trichoderma viride. Appl. Biochem. Microbiol. 47: 53–58.
Kahar, P., T. Iwata, J. Hiraki, E. Y. Park, and M. Okabe (2001) Enhancement of ε-polylysine production by Streptomyces albulus strain 410 using pH control. J. Biosci. Bioeng. 91: 190–194.
Kito, M., R. Takimoto, Y. Onji, T. Yoshida, and T. Nagasawa (2002) Purification and characterization of an ε-poly-L-lysine-degrading enzyme from the e-poly-L-lysine-tolerant Chryseobacterium sp. OJ7. J. Biosci. Bioeng. 96: 92–104.
Helena, B., H. G. Nimmo, I. S. Hunter, and J. R. Coggins (1993) Phosphoenolpyruvate carboxylase from Streptomyces coelicolor A3(2): purification of the enzyme, cloning of the ppc gene and over-expression of the protein in a streptomycete. Biochem. J. 293: 131–136.
Hosoya, Y., S. Okamoto, H. Muramatsu, and K. Ochi (1998) Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob. Agents Chemother. 42: 2041–2047.
Wang, G., T. Hosaka, and K. Ochi (2008) Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl. Environ. Microbol. 74: 2834–2840.
Acknowledgments
This work was financially supported by Shandong Natural Science Foundation (ZR2019BC044), and open project from the Key Laboratory of Industrial Biotechnology, Ministry of Education, Jiangnan University (KLIB-KF202007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Neither ethical approval nor informed consent was required for this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Conflict of Interest
The authors have no financial conflicts of interest to declare.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Li, S., Wang, L., Wang, N. et al. Combining Genome Shuffling with Streptomycin Resistance to Improve Poly-γ-L-diaminobutanoic Acid Production in Bacillus pumilus. Biotechnol Bioproc E 26, 630–640 (2021). https://doi.org/10.1007/s12257-020-0320-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-020-0320-2