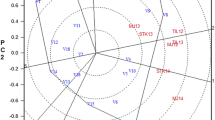
Abstract
Genotype x environment interactions (GEI) slows the genetic progress in breeding through reduced selection gains. The additive main effects and multiplicative interaction (AMMI) analysis and genotype main effect and genotype x environment interaction (GGE) biplot analysis are widely used to measure stability of yield and its components. The objective of this study was to estimate the magnitude of GEI for potato tuber yield and bacterial wilt resistance and to identify the most discriminating and representative environments for potato testing in Kenya. The study was conducted in four environments. Forty eight potato families were evaluated using a 6 × 8 alpha lattice design replicated three times. Data on days from planting to onset of wilting, area under the disease progress curve, total tuber weight (t ha−1), total tuber numbers/hectare, proportion of ware sized tubers, proportion of symptomatic tubers based on weight, proportion of symptomatic tubers based on tuber numbers, and latent infection of the tubers were subjected to combined analysis of variance in order identify crosses that were resistant to bacterial wilt. Data on tuber yields were analysed using AMMI and GGE biplot methods in order to identify the highest yielding and most stable family as well as the most discriminating and yet representative test environment. Family 20 was closest to the ideal genotype; it was the highest yielding (104.686 t ha−1) and most stable; it was followed by family 43. The environment ENVI 1 was the closest to ideal environment and therefore the most desirable of the four test environments.
Resumen
Las interacciones genotipo-medio ambiente (GEI) hacen más lento el progreso genético en mejoramiento mediante la reducción de logros por selección. El análisis de los principales efectos aditivos y la interacción multiplicativa (AMMI) y el principal efecto del genotipo, y el análisis de multivariables superpuestas (biplot) de la interacción genotipo-medio ambiente (GGE), se usan ampliamente para medir la estabilidad del rendimiento y sus componentes. El objetivo de este estudio fue estimar la magnitud de GEI para el rendimiento del tubérculo en papa y la resistencia a la marchitez bacteriana, y para identificar los ambientes más discriminadores y representativos para pruebas de papa en Kenia. El estudio se condujo en cuatro ambientes. Se evaluaron 48 familias de papa utilizando un diseño de alfa látice de 6 × 8 con tres repeticiones. Los datos de días de siembra hasta el establecimiento de la marchitez, del área bajo la curva de progreso de la enfermedad, del peso total del tubérculo (t ha−1), del número total de tubérculos por hectárea, de la proporción de tubérculos de tamaño comercial, de la proporción de tubérculos sintomáticos con base en el peso, de la proporción de tubérculos sintomáticos con base en números y de la infección latente de tubérculos, estuvieron sujetos a un análisis de varianza combinado a fin de identificar cruzas que fueran resistentes a la marchitez bacteriana. Se analizaron los datos de rendimiento de tubérculo usando los métodos biplot AMMI y GGE para identificar la familia más estable y de más altos rendimientos, así como del ambiente probado más discriminador y a la vez representativo. La familia 20 fue la más cercana al genotipo ideal; fue la de más alto rendimiento (104.686 t ha−1) y la más estable; seguida por la familia 43. El ambiente ENVI 1 fue el más cercano al ideal, y por lo tanto el más deseable de los cuatro ambientes probados.





Similar content being viewed by others
References
Bernardo, R. 2002. Breeding for quantitative traits in plants. Woodbury: Stemma Press.
Buddenhagen, I., and A. Kelman. 1964. Biological and physiological aspects of bacterial wilt caused by P. solanacearum. Annual Review of Plant Pathology 2: 203–230.
CIP. 2007. Procedures for standard evaluation trials of advanced potato clones. An International Cooperators’ Guide. Lima: Centro Internacional de la Papa.
Cooper, M., and J.H. Delacy. 1994. Relationships among analytical methods used to study genotypic variation and genotype - by - environment interaction in plant breeding multi-environment experiments. Theoretical and Applied Genetics 88: 561–572.
Cooper, M., R.M. Stuker, I.H. Delacy, and B.D. Harch. 1997. Wheat breeding nurseries, target environments, and indirect selection for grain yield. Crop Science 37: 1168–1176.
Crossa, J. 1990. Statistical analyses of multilocation trials. Advances in Agronomy 44: 55–85.
Crossa, J., P.L. Cornelius, K. Sayre, and J.I.R. Ortiz-Monasterio. 1995. A shifted multiplicative model fusion method for grou** environments without cultivar rank change. Crop Science 35: 54–62.
Dabholkar, A.R. 1992. Elements of biometrical genetics, 1st ed. New Delhi: Concept Publishing Company.
Falconer, D.S., and T.F.C. Mackay. 1996. Introduction to quantitative genetics, 4th ed. Harlow: Pearson Prentice Hall.
Fox, P.N., J. Crossa, and I. Romagosa. 1997. Multi-environment testing and genotype x environment interaction. In Statistical methods for plant variety evaluation, ed. R.A. Kempton and P.N. Fox, 117–138. London: Chapman and Hall.
French, E.R., and L.D. Lindo. 1982. Resistance to Pseudomonas solanacearum in potato: SPECIFICITY and temperature sensitivity. Phytopathology 72: 1408–1412.
Gauch, H.G., and R.W. Zobel. 1988. Predictive and postdictive success of statistical analyses of yield trials. Theoretical and Applied Genetics 76: 1–10.
Gauch, H.G., and R.W. Zobel. 1997. Identifying mega-environments and targeting genotypes. Crop Science 37: 311–326.
Harris, O.C. 1976. Bacterial wilt in Kenya with particular reference to potatoes. In Proceedings of the first international planning conference and workshop on the ecology and control of bacterial wilt caused by Pseudomonas solanacearum, Raleigh, North Carolina.18-24 January 1976, ed. L. Sequeira and A. Kelman, 84–88. Berlin: Springer-Verlag.
Haverkort, A.J., M. van de Waart, and K.B.A. Bodlaender. 1990. The effect of early drought stress on numbers of tubers and stolons of potato in controlled and field conditions. Potato Research 33: 89–96.
Jaetzold, R., H. Schmidt, B. Hornetz, and C. Shisanya. 2006. Farm Management Handbook of Kenya. Natural conditions and farm management information. Part B. Central Kenya. Subpart B1. South Rift, vol. II, 2nd ed. Nairobi: Ministry of Agriculture.
Kang, M.S. 1993. Simultaneous selection for yield and stability in crop performance trials: consequences for growers. Agronomy Journal 85: 754–757.
KARI. 2008. Production of food (ware) potatoes. KARI information brochure. Nairobi: Kenya Agricultural Research Institute.
Martin, C., and E.R. French. 1985. Bacterial wilt of potatoes caused by Pseudomonas solanacearum. Technical Information Bulletin 13: 1–6. Centro Internacional de la Papa, Lima, Peru.
Mather, K., and J.L. **ks. 1982. Biometrical genetics, 3rd ed. London: Chapman and Hall.
Muthoni, J., H. Shimelis, R. Melis, and Z.M. Kinyua. 2014. Response of potato genotypes to bacterial wilt caused by Ralstonia solanacearum (Smith)(Yabuuchi et al.) In the tropical highlands. American Journal of Potato Research 91: 215–232. doi:10.1007/s12230-013-9340-1.
Payne, R.W., D.A. Murray, S. A. Harding, D. B. Baird, and D. M. Soutar. 2011. GenStat for Windows 14th ed. Introduction. VSN International, Hemel Hempstead.
Priou, S., L. Gutarra, and P. Aley. 1999. Highly sensitive detection of Ralstonia solanacearum in latently infected potato tubers by post-enrichment ELISA on nitrocellulose membrane. EPPO/OEPP Bulletin 29: 117–125.
Sas, I. 2003. SAS user’s guide. Cary: In Statistics.
SPSS Inc. 2009. Statistical Package for Social Scientists. SPSS for Windows Release 18.0. 2009. SPSS Inc. 2009. Chicago, IL, www.spss.com.
Tung, P.X. 1992. Genetic variation for bacterial wilt resistance in a population of tetraploid potato. Euphytica 61: 73–80.
Tung, P.X., E.T. Rasco, P.V. Zaag, and P. Schmiediche. 1990. Resistance to Pseudomonas solanacearum in the potato: I. Effects of sources of resistance and adaptation. Euphytica 45: 203–210.
Tung, P.X., J.G.T. Hermsen, P.v.d. Zaag, and P. Schmiediche. 1992a. Effects of resistance genes, heat tolerance genes and cytoplasms on expression of resistance to Pseudomonas solanacearum (E.F. Smith) in potato. Euphytica 60: 127–138.
Tung, P.X., J.G.T. Hermsen, P.v.d. Zaag, and P. Schmiediche. 1992b. Effects of heat tolerance on expression of resistance to Pseudomonas solanacearum E. F. Smith in potato. Potato Research 35: 321–328.
Tung, P.X., J.G. Hermsen, P.Z. Vander, and P.E. Schmiediche. 2006. Inheritance of resistance to Pseudomonas solanacearum in tetraploid potato. Plant Breeding 111: 23–30.
UNESCO. 1977. FAO-unesco soil map of the world. Vol. VI. Africa, vol. VI. Paris: UNESCO.
Yan, W. 2002. Singular-value partitioning in biplot analysis of multi-environment trial data. Agronomy Journal 94: 990–996.
Yan, W., and M.S. Kang. 2003. GGE biplot analysis: a graphical tool for breeders, geneticists, and agronomists. New York: RCR Press.
Yan, W., and N.A. Tinker. 2006. Biplot analysis of multi-environment trial data; principles and application. Canadian Journal of Plant Science 86: 623–645.
Yan, W., L.A. Hunt, Q. Sheng, and Z. Szlavnics. 2000. Cultivar evaluation and mega-environment investigation based on the GGE Biplot. Crop Science 40: 597–605.
Yan, W., P.L. Cornelius, J. Crossa, and L.A. Hunt. 2001. Two types of GGE Biplots for analyzing multi-environment trial data. Crop Science 41: 656–663.
Yan, W., B.M. Kang, S. Woods, and P.L. Cornelius. 2007. GGE biplot vs AMMI analysis of genotype-by-genotype environment data. Crop Science 47: 643–655.
Acknowledgments
Due thanks go to the Alliance for a Green Revolution in Africa (AGRA) for funding this research and Kenya Agricultural Research Institute (KARI) management for granting study leave to the first author.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Muthoni, J., Shimelis, H. & Melis, R. Genotype x Environment Interaction and Stability of Potato Tuber Yield and Bacterial Wilt Resistance in Kenya. Am. J. Potato Res. 92, 367–378 (2015). https://doi.org/10.1007/s12230-015-9442-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12230-015-9442-z