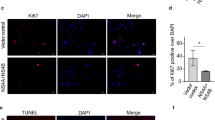
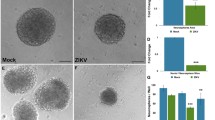
Abstract
Brain abnormalities and congenital malformations have been linked to the circulating strain of Zika virus (ZIKV) in Brazil since 2016 during the microcephaly outbreak; however, the molecular mechanisms behind several of these alterations and differential viral molecular targets have not been fully elucidated. Here we explore the proteomic alterations induced by ZIKV by comparing the Brazilian (Br ZIKV) and the African (MR766) viral strains, in addition to comparing them to the molecular responses to the Dengue virus type 2 (DENV). Neural stem cells (NSCs) derived from induced pluripotent stem (iPSCs) were cultured both as monolayers and in suspension (resulting in neurospheres), which were then infected with ZIKV (Br ZIKV or ZIKV MR766) or DENV to assess alterations within neural cells. Large-scale proteomic analyses allowed the comparison not only between viral strains but also regarding the two- and three-dimensional cellular models of neural cells derived from iPSCs, and the effects on their interaction. Altered pathways and biological processes were observed related to cell death, cell cycle dysregulation, and neurogenesis. These results reinforce already published data and provide further information regarding the biological alterations induced by ZIKV and DENV in neural cells.






Similar content being viewed by others
Data Availability
The proteomic datasets generated for this study can be found in the PRIDE proteomics data repository (https://www.ebi.ac.uk/pride/archive/) with the accession numbers PXD026825 and PXD026909.
References
World Health Organization (2019) Zika epidemiology update. 1–14.
Brasil P, Pereira JP, Moreira ME et al (2016) Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med 375:2321–2334. https://doi.org/10.1056/NEJMoa1602412
de Araujo TVB, Rodrigues LC, de Alencar **menes RA et al (2016) Association between Zika virus infection and microcephaly in Brazil, January to May, 2016: preliminary report of a case-control study. Lancet Infect Dis 16:1356–1363. https://doi.org/10.1016/S1473-3099(16)30318-8
Golubeva VA, Nepomuceno TC, de Gregoriis G et al (2020) Network of interactions between ZIKA virus non-structural proteins and human host proteins. Cells 9:153. https://doi.org/10.3390/cells9010153
Liu Z-Y, Shi W-F, Qin C-F (2019) The evolution of Zika virus from Asia to the Americas. Nat Rev Microbiol 17:131–139. https://doi.org/10.1038/s41579-018-0134-9
Musso D, Ko AI, Baud D (2019) Zika virus infection — after the pandemic. N Engl J Med 381:1444–1457. https://doi.org/10.1056/NEJMra1808246
Pan American Health Organization (2018) Zika cases and congenital syndrome associated with Zika virus reported by countries and territories in the Americas, 2015–2018: cumulative cases. 1–1.
Bayer A, Lennemann NJ, Ouyang Y et al 2016 Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection Cell Host Microbe 1 9 https://doi.org/10.1016/j.chom.2016.03.008
Calvet G, Aguiar RS, Melo ASO et al 2016 Articles Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study Lancet Infect Dis 1 8 https://doi.org/10.1016/S1473-3099(16)00095-5
Woods CG, Bond J, Enard W (2005) Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. The American Journal of Human Genetics 76:717–728. https://doi.org/10.1086/429930
Woods CG, Parker A (2013) Investigating microcephaly. Arch Dis Child 98:707–713. https://doi.org/10.1136/archdischild-2012-302882
Ming G-L, Tang H, Song H (2016) Advances in Zika virus research: stem cell models, challenges, and opportunities. Stem Cell 19:690–702. https://doi.org/10.1016/j.stem.2016.11.014
McGrath EL, Rossi SL, Gao J et al (2017) Differential responses of human fetal brain neural stem cells to Zika virus infection. Stem Cell Reports 8:715–727. https://doi.org/10.1016/j.stemcr.2017.01.008
Martines RB, Bhatnagar J, Keating MK et al (2016) Notes from the field: evidence of Zika virus infection in brain and placental tissues from two congenitally infected newborns and two fetal losses — Brazil, 2015. MMWR Morb Mortal Wkly Rep 65:159–160. https://doi.org/10.15585/mmwr.mm6506e1
Pierson TC, Diamond MS (2018) The emergence of Zika virus and its new clinical syndromes. Nature 560:573–581. https://doi.org/10.1038/s41586-018-0446-y
de Miranda-Filho D, B, Martelli CMT, **menes RA de A, et al (2016) Initial description of the presumed congenital Zika syndrome. Am J Public Health 106:598–600. https://doi.org/10.2105/AJPH.2016.303115
Cugola FR, Fernandes IR, Russo FB et al (2016) The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534:267–271. https://doi.org/10.1038/nature18296
Dang J, Tiwari SK, Lichinchi G et al (2016) Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Stem Cell 19:258–265. https://doi.org/10.1016/j.stem.2016.04.014
Garcez PP, Loiola EC, Madeiro da Costa R et al (2016) Zika virus impairs growth in human neurospheres and brain organoids. Science 352:816–818. https://doi.org/10.1126/science.aaf6116
Li C, Xu D, Ye Q et al (2016) Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. https://doi.org/10.1016/j.stem.2016.04.017
Qian X, Nguyen HN, Song MM et al (2016) Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165:1238–1254. https://doi.org/10.1016/j.cell.2016.04.032
Tang H, Hammack C, Ogden SC et al (2016) Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 18:1–22. https://doi.org/10.1016/j.stem.2016.02.016
Ledur PF, Karmirian K, Pedrosa CDSG et al (2020) Zika virus infection leads to mitochondrial failure, oxidative stress and DNA damage in human iPSC-derived astrocytes. Sci Rep 10:1218–1314. https://doi.org/10.1038/s41598-020-57914-x
Garcez PP, Nascimento JM, de Vasconcelos JM, et al (2017) Zika virus disrupts molecular fingerprinting of human neurospheres. Sci Rep 7:40780 EP –. https://doi.org/10.1038/srep40780
Lancaster MA, Renner M, Martin C-A et al (2013) Cerebral organoids model human brain development and microcephaly. Nature 501:373–379. https://doi.org/10.1038/nature12517
Zhang F, Hammack C, Ogden SC et al (2016) Molecular signatures associated with ZIKV exposure in human cortical neural progenitors. Nucleic Acids Res 44:8610–8620. https://doi.org/10.1093/nar/gkw765
Esser-Nobis K, Aarreberg LD, Roby JA et al (2019) Comparative analysis of African and Asian lineage-derived Zika virus strains reveals differences in activation of and sensitivity to antiviral innate immunity. J Virol 93:e00640-e719. https://doi.org/10.1128/JVI.00640-19
Willard KA, Demakovsky L, Tesla B et al (2017) Zika virus exhibits lineage-specific phenotypes in cell culture, in Aedes aegypti mosquitoes, and in an embryo model. Viruses 9:383. https://doi.org/10.3390/v9120383
Aubry F, Jacobs S, Darmuzey M et al (2021) Recent African strains of Zika virus display higher transmissibility and fetal pathogenicity than Asian strains. Nat Comms 12:916–1014. https://doi.org/10.1038/s41467-021-21199-z
Annamalai AS, Pattnaik A, Sahoo BR et al (2017) Zika virus encoding nonglycosylated envelope protein is attenuated and defective in neuroinvasion. J Virol. https://doi.org/10.1128/JVI.01348-17
Anfasa F, Siegers JY, van der Kroeg M, et al (2017) Phenotypic differences between Asian and African lineage Zika viruses in human neural progenitor cells. mSphere
Sirohi D, Chen Z, Sun L, et al. (2016) The 3.8 Å resolution cryo-EM structure of Zika virus. Science. https://doi.org/10.1126/science.aaf5316
Zhang X, Ge P, Yu X et al (2013) Cryo-EM structure of the mature Dengue virus at 3.5-Å resolution. Nat Struct Mol Biol 20:105–110
Carod-Artal FJ, Wichmann O, Farrar J, Gascón J (2013) Neurological complications of Dengue virus infection. Lancet Neurol 12:906–919. https://doi.org/10.1016/S1474-4422(13)70150-9
Shevchenko A, Tomas H, Havlis J et al (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. https://doi.org/10.1038/nprot.2006.468
Cassoli JS, Brandao-Teles C, Santana AG et al (2017) Ion mobility-enhanced data-independent acquisitions enable a deep proteomic landscape of oligodendrocytes. Proteomics 17:1700209. https://doi.org/10.1002/pmic.201700209
Zhou Y, Zhou B, Pache L et al (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Comms 10:1523. https://doi.org/10.1002/sim.4780090710
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4:2–27. https://doi.org/10.1186/1471-2105-4-2
Szklarczyk D, Gable AL, Lyon D et al (2018) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Jassal B, Matthews L, Viteri G et al (2020) The reactome pathway knowledgebase. Nucleic Acids Res 48:D498–D503. https://doi.org/10.1093/nar/gkz1031
Giurgiu M, Reinhard J, Brauner B et al (2019) CORUM: the comprehensive resource of mammalian protein complexes—2019. Nucleic Acids Res 47:D559–D563. https://doi.org/10.1093/nar/gky973
Kanehisa M, Furumichi M, Tanabe M et al (2016) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. https://doi.org/10.1016/j.febslet.2013.06.026
Allgoewer K, Maity S, Zhao A et al (2021) New proteomic signatures to distinguish between Zika and Dengue infections. Mol Cell Proteomics 20:100052. https://doi.org/10.1016/j.mcpro.2021.100052
Gabriel E, Ramani A, Karow U et al (2017) Recent Zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell 20:397-406.e5. https://doi.org/10.1016/j.stem.2016.12.005
Chen LS, Shi SJ, Zou PS, et al (2016) Identification of novel DYNC2H1 mutations associated with short rib-polydactyly syndrome type III using next-generation panel sequencing. Genetics and molecular research : GMR. https://doi.org/10.4238/gmr.15028134
Fujita A, Higashijima T, Shirozu H et al (2019) Pathogenic variants of DYNC2H1, KIAA0556, and PTPN11 associated with hypothalamic hamartoma. Neurology 93:e237–e251. https://doi.org/10.1212/WNL.0000000000007774
Li G-H, Ning Z-J, Liu Y-M, Li X-H (2017) Neurological manifestations of Dengue infection. Front Cell Infect Microbiol 7:449. https://doi.org/10.3389/fcimb.2017.00449
Buttitta LA, Edgar BA (2007) How size is controlled: from Hippos to Yorkies. Nat Cell Biol 9:1225–1227
Garcia GJ, Paul S, Beshara S et al (2020) Hippo signaling pathway has a critical role in Zika virus replication and in the pathogenesis of neuroinflammation. Am J Pathol 190:844–861. https://doi.org/10.1016/j.ajpath.2019.12.005
Mo J-S, Park HW, Guan K-L (2014) The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep 15:642–656. https://doi.org/10.15252/embr.201438638
Giglione C, Fieulaine S, Meinnel T (2015) N-terminal protein modifications: bringing back into play the ribosome. Biochimie 114:134–146. https://doi.org/10.1016/j.biochi.2014.11.008
Thinon E, Serwa RA, Broncel M et al (2014) Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat Comms 5:4919–4919. https://doi.org/10.1038/ncomms5919
Suwanmanee S, Mahakhunkijcharoen Y, Ampawong S et al (2019) Inhibition of N-myristoyltransferase1 affects Dengue virus replication. MicrobiologyOpen 8:e00831. https://doi.org/10.1002/mbo3.831
Deans AJ, West SC (2011) DNA interstrand crosslink repair and cancer. Nat Rev Cancer 11:467–480
Tiwari SK, Dang JW, Lin N et al (2020) Zika virus depletes neural stem cells and evades selective autophagy by suppressing the Fanconi anemia protein FANCC. EMBO Rep 21:e49183. https://doi.org/10.15252/embr.201949183
Gleeson JG, Minnerath SR, Fox JW et al (1999) Characterization of mutations in the gene doublecortin in patients with double cortex syndrome. Ann Neurol 45:146–153. https://doi.org/10.1002/1531-8249(199902)45:2%3c146::aid-ana3%3e3.0.co;2-n
Jiang X, Dong X, Li S-H et al (2018) Proteomic analysis of Zika virus infected primary human fetal neural progenitors suggests a role for doublecortin in the pathological consequences of infection in the cortex. Front Microbiol 9:1067–1067. https://doi.org/10.3389/fmicb.2018.01067
Scaturro P, Stukalov A, Haas DA et al (2018) An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature 561:253–257. https://doi.org/10.1038/s41586-018-0484-5
Coyaud E, Ranadheera C, Cheng D et al (2018) Global interactomics uncovers extensive organellar targeting by Zika virus. Mol Cell Proteomics 17:2242–2255. https://doi.org/10.1074/mcp.TIR118.000800
Bartuzi P, Billadeau DD, Favier R et al (2016) CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nat Comms 7:10961–11011. https://doi.org/10.1038/ncomms10961
Hashimoto Y, Sheng X, Murray-Nerger LA, Cristea IM (2020) Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection. Nat Comms 11:806
Gomez TS, Gorman JA, Artal-Martinez de Narvajas A et al (2012) Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol Biol Cell 23:3215–3228. https://doi.org/10.1091/mbc.e12-02-0101
De Maio FA, Risso G, Iglesias NG et al (2016) The Dengue virus NS5 protein intrudes in the cellular spliceosome and modulates splicing. PLoS Pathog 12:e1005841. https://doi.org/10.1371/journal.ppat.1005841
Martinez O, Goud B (1998) Rab proteins. Biochem Biophys Acta 1404:101–112. https://doi.org/10.1016/s0167-4889(98)00050-0
Wanschers BFJ, van de Vorstenbosch R, Schlager MA et al (2007) A role for the Rab6B Bicaudal-D1 interaction in retrograde transport in neuronal cells. Exp Cell Res 313:3408–3420. https://doi.org/10.1016/j.yexcr.2007.05.032
Spearman P (2018) Viral interactions with host cell Rab GTPases. Small GTPases 9:192–201. https://doi.org/10.1080/21541248.2017.1346552
Scrima A, Thomas C, Deaconescu D, Wittinghofer A (2008) The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues. EMBO J 27:1145–1153. https://doi.org/10.1038/emboj.2008.30
Niemann JH, Du C, Morlot S et al (2020) De novo missense variants in the RAP1B gene identified in two patients with syndromic thrombocytopenia. Clin Genet 98:374–378. https://doi.org/10.1111/cge.13807
Colledge M, Scott JD (1999) AKAPs: from structure to function. Trends Cell Biol 9:216–221. https://doi.org/10.1016/s0962-8924(99)01558-5
Larocca MC, Shanks RA, Tian L et al (2004) AKAP350 interaction with cdc42 interacting protein 4 at the Golgi apparatus. Mol Biol Cell 15:2771–2781. https://doi.org/10.1091/mbc.e03-10-0757
Reid CR, Airo AM, Hobman TC (2015) The virus-host interplay: biogenesis of +RNA replication complexes. Viruses 7:4385–4413. https://doi.org/10.3390/v7082825
Sirohi D, Kuhn RJ (2017) Zika virus structure, maturation, and receptors. J INFECT DIS 216:S935–S944. https://doi.org/10.1093/infdis/jix515
Garcez PP, Diaz-Alonso J, Crespo-Enriquez I et al (2015) Cenpj/CPAP regulates progenitor divisions and neuronal migration in the cerebral cortex downstream of Ascl1. Nat Comms 6:1–14. https://doi.org/10.1038/ncomms7474
Sanchez EL, Lagunoff M (2015) Viral activation of cellular metabolism. Virology 479–480:609–618. https://doi.org/10.1016/j.virol.2015.02.038
Gilbert Jaramillo J, Garcez P, James W et al (2019) The potential contribution of impaired brain glucose metabolism to congenital Zika syndrome. J Anat 235:468–480. https://doi.org/10.1111/joa.12959
Borrell V, Cárdenas A, Ciceri G et al (2012) Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron 76:338–352. https://doi.org/10.1016/j.neuron.2012.08.003
Blockus H, Chédotal A (2016) Slit-Robo signaling Development 143:3037–3044. https://doi.org/10.1242/dev.132829
Dykes IM, Lanier J, Raisa Eng S, Turner EE (2010) Brn3a regulates neuronal subtype specification in the trigeminal ganglion by promoting Runx expression during sensory differentiation. Neural Dev 5:3–18. https://doi.org/10.1186/1749-8104-5-3
Wang JW, Stifani S (2017) Roles of Runx genes in nervous system development. Adv Exp Med Biol 962:103–116. https://doi.org/10.1007/978-981-10-3233-2_8
Brault J-B, Khou C, Basset J et al (2016) EBioMedicine EBIOM 10:71–76. https://doi.org/10.1016/j.ebiom.2016.07.018
Ferraris P, Cochet M, Hamel R et al (2019) Zika virus differentially infects human neural progenitor cells according to their state of differentiation and dysregulates neurogenesis through the Notch pathway. Emerg Microbes Infect 8:1003–1016. https://doi.org/10.1080/22221751.2019.1637283
Costa VV, Del Sarto Juliana L, Rocha RF, et al (2017) N-methyl-d-aspartate (NMDA) receptor blockade prevents neuronal death induced by Zika virus infection. mBio 8:e00350–17. https://doi.org/10.1128/mBio.00350-17
Serras F, Morey M, Alsina B et al (2001) The Drosophila selenophosphate synthetase (selD) gene is required for development and cell proliferation. BioFactors (Oxford, England) 14:143–149. https://doi.org/10.1002/biof.5520140119
Acknowledgements
The authors thank Gabriela Vitoria, Ismael Gomes, Paulo Baldasso, and Erick Loiola for their excellent technical support and Bradley Smith, MSc for the critical comments and English review support during the process.
Funding
Financial support was provided by the São Paulo Research Foundation (D. G. J., J. M. N., G. S. Z., C. B. T., J. S. C., A. S. L. M. A. and D. M. S. are supported by FAPESP grant numbers 2014/14881-1, 2014/21035-0, 2017/25055-3, 2018/25439–9, 2018/14666–4, 2020/10282-7, 2017/25588–1, 2019/00098-7), and the National Council of Scientific and Technological Development (CNPq), in addition to intramural grants from D’Or Institute for Research and Education.
Author information
Authors and Affiliations
Contributions
J. M. N., P. P. G., S. K. R., and D. M. S. conceived and designed the study. D. J. G. and J. M. N. performed in silico proteomic analyses. J. M. N. and J. S. C. performed the mass spectrometry experiments. G. S. Z. performed the pathway analysis and interpretation. P. P. G., C. S. G. P., K. K., J. A. S., A. S. L. M. A., C. B. T., L. M. H., G. F. S., S. P. M., and F. C. cultured iPS cells and/or virus strains, performed cell-based assays and/or infection, and contributed to discussion. A. T. and J. L. P. M. performed the data interpretation. D. J. G. and J. M. N. interpreted the data, wrote, edited, and revised the manuscript. D. M. S. and S. K. R. coordinated the study. All authors contributed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nascimento, J.M., Gouvêa-Junqueira, D., Zuccoli, G.S. et al. Zika Virus Strains and Dengue Virus Induce Distinct Proteomic Changes in Neural Stem Cells and Neurospheres. Mol Neurobiol 59, 5549–5563 (2022). https://doi.org/10.1007/s12035-022-02922-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-02922-3