Abstract
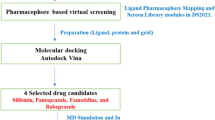
Recent studies highlight the great potential impact of HDAC inhibitors (HDACis) in suppressing TNBC, even though clinical trials including a single HDACis demonstrated unsatisfactory outcomes against TNBC. New compounds created to achieve isoform selectivity and/or a polypharmacological HDAC strategy have also produced interesting results. The current study discusses the HDACis pharmacophoric models and the structural alterations that produced drugs with strong inhibitory effects on TNBC progression. With more than 2 million new cases reported in 2018, breast cancer—the most common cancer among women worldwide—poses a significant financial burden on an already deteriorating public health system. Due to a lack of therapies being developed for triple-negative breast cancers and the development of resistance to the current treatment options, it is imperative to plan novel therapeutics in order to bring new medications to the pipeline. Additionally, HDACs deacetylate a large number of nonhistone cellular substrates that control a variety of biological processes, such as the beginning and development of cancer. The significance of HDACs in cancer and the therapeutic potential of HDAC inhibitor. Furthermore, we also reported molecular docking study with four HDAC inhibitors and performed molecular dynamic stimulation of the best dock score compound. Among the four ligands belinostat compound showed best binding affinity with histone deacetylase protein which was -8.7 kJ/mol. It also formed five conventional hydrogen bond with Gly 841, His 669, His 670, pro 809, and His 709 amino acid residues.










Similar content being viewed by others
Data availability
No data were used for the research described in the article.
References
Duranti S, Fabi A, Filetti M, Falcone R, Lombardi P, Daniele G, Franceschini G, Carbognin L, Palazzo A, Garganese G, et al. Breast cancer drug approvals issued by EMA: a review of clinical trials. Cancers. 2021;13:5198. https://doi.org/10.3390/cancers13205198.
Zolota V, Tzelepi V, Piperigkou Z, Kourea H, Papakonstantinou E, Argentou MI, Karamanos NK. Epigenetic alterations in triple-negative breast cancer—the critical role of extracellular matrix. Cancers. 2021;13:713. https://doi.org/10.3390/cancers13040713.
Yoshida M, Kudo N, Kosono S, Ito A. Chemical and structural biology of protein lysine deacetylases. Proc Jpn Acad Ser B. 2017;93:297–321. https://doi.org/10.2183/pjab.93.019.
Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–52. https://doi.org/10.1038/38664.
Jenke R, Reßing N, Hansen F, Aigner A, Büch T. Anticancer therapy with HDAC inhibitors: mechanism-based combination strategies and future perspectives. Cancers. 2021;13:634. https://doi.org/10.3390/cancers13040634.
Terranova-Barberio M, Thomas S, Ali N, Pawlowska N, Park J, Krings G, Rosenblum MD, Budillon A, Munster PN. HDAC inhibition potentiates immunotherapy in triple negative breast cancer. Oncotarget. 2017;8:114156–72. https://doi.org/10.18632/oncotarget.23169.
Maiti A, Qi Q, Peng X, Yan L, Takabe K, Hait NC. Class I histone deacetylase inhibitor suppresses vasculogenic mimicry by enhancing the expression of tumor suppressor and anti-angiogenesis genes in aggressive human TNBC cells. Int J Oncol. 2019;55:116–30. https://doi.org/10.3892/ijo.2019.4796.
Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci USA. 2004;101:15064–9. https://doi.org/10.1073/pnas.0404603101.
Bressi JC, Jennings AJ, Skene R, Wu Y, Melkus R, De Jong R, O’Connell S, Grimshaw CE, Navre M, Gangloff AR. Exploration of the HDAC2 foot pocket: synthesis and SAR of substituted N-(2-aminophenyl)benzamides. Bioorg Med Chem Lett. 2010;20:3142–5. https://doi.org/10.1016/j.bmcl.2010.03.091.
Roche J, Bertrand P. Inside HDACs with more selective HDAC inhibitors. Eur J Med Chem. 2016;121:451–83. https://doi.org/10.1016/j.ejmech.2016.05.047.
Ho TCS, Chan AHY, Ganesan A. Thirty years of HDAC inhibitors: 2020 insight and hindsight. J Med Chem. 2020;63:12460–84. https://doi.org/10.1021/acs.jmedchem.0c00830.
Connolly RM, Rudek MA, Piekarz R. Entinostat: a promising treatment option for patients with advanced breast cancer. Future Oncol. 2017;13:1137–48. https://doi.org/10.2217/fon-2016-0526.
Zhang MC, Fang Y, Wang L, Cheng S, Fu D, He Y, Zhao Y, Wang CF, Jiang XF, Song Q, et al. Clinical efficacy and molecular biomarkers in a phase II study of tucidinostat plus R-CHOP in elderly patients with newly diagnosed diffuse large B-cell lymphoma. Clin Epigenet. 2020;12:160. https://doi.org/10.1186/s13148-020-00948-9.
Santo L, Hideshima T, Kung A, Tseng JC, Tamang D, Yang M, Jarpe M, Van Duzer JH, Mazitschek R, Ogier WC, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood. 2012;119:2579–89. https://doi.org/10.1182/blood-2011-10-387365.
Huang P, Almeciga-Pinto I, Jarpe M, Van Duzer JH, Mazitschek R, Yang M, Jones SS, Quayle SN. Selective HDAC inhibition by ACY-241 enhances the activity of paclitaxel in solid tumor models. Oncotarget. 2017;8:2694–707. https://doi.org/10.18632/oncotarget.13738.
Nawar N, Bukhari S, Adile AA, Suk Y, Manaswiyoungkul P, Toutah K, Olaoye OO, Raouf YS, Sedighi A, Garcha HK, et al. Discovery of HDAC6-selective inhibitor NN-390 with in vitro efficacy in group 3 medulloblastoma. J Med Chem. 2022;65:3193–217. https://doi.org/10.1021/acs.jmedchem.1c01585.
Li S, Zhao C, Zhang G, Xu Q, Liu Q, Zhao W, Chou CJ, Zhang Y. Development of selective HDAC6 inhibitors with in vitro and in vivo anti-multiple myeloma activity. Bioorg Chem. 2021;116:105278. https://doi.org/10.1016/j.bioorg.2021.105278.
Depetter Y, Geurs S, De Vreese R, Goethals S, Vandoorn E, Laevens A, Steenbrugge J, Meyer E, De Tullio P, Bracke M, et al. Selective pharmacological inhibitors of HDAC6 reveal biochemical activity but functional tolerance in cancer models. Int J Cancer. 2019;145:735–47. https://doi.org/10.1002/ijc.32169.
Chan TS, Tse E, Kwong YL. Chidamide in the treatment of peripheral T-cell lymphoma. OncoTargets Ther. 2017;10:347–52. https://doi.org/10.2147/OTT.S93528.
Millán-Zambrano G, Burton A, Bannister AJ, et al. Histone post-translational modifications — cause and consequence of genome function. Nat Rev Genet. 2022;23:563–80.
Wu S, Luo Z, Yu PJ, **e H, He YW. Suberoylanilide hydroxamic acid (SAHA) promotes the epithelial mesenchymal transition of triple negative breast cancer cells via HDAC8/FOXA1 signals. Biol Chem. 2016;397:75–83. https://doi.org/10.1515/hsz-2015-0215.
Woo YM. Epigenetic regulation in cystogenesis. Adv Exp Med Biol. 2016;933:59–68.
Li H, Gong Y, Zhong Q. In vivo anticancer potential of hydroxamic acid derivatives. Curr Top Med Chem. 2021;21:1737–55. https://doi.org/10.2174/1568026621666210813105240.
Carlisi D, Lauricella M, D’Anneo A, Buttitta G, Emanuele S, di Fiore R, Martinez R, Rolfo C, Vento R, Tesoriere G. The synergistic effect of SAHA and parthenolide in MDA-MB231 breast cancer cells. J Cell Physiol. 2015;230:1276–89. https://doi.org/10.1002/jcp.24863.
Chiu HW, Yeh YL, Wang YC, Huang WJ, Chen YA, Chiou YS, Ho SY, Lin P, Wang YJ. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, enhances radiosensitivity and suppresses lung metastasis in breast cancer in vitro and in vivo. PLoS ONE. 2013;8:e76340. https://doi.org/10.1371/journal.pone.0076340.
Sabnis GJ, Goloubeva O, Chumsri S, Nguyen N, Sukumar S, Brodie AMH. Functional activation of the estrogen receptor-α and aromatase by the HDAC inhibitor entinostat sensitizes ER-negative tumors to letrozole. Cancer Res. 2011;71:1893–903. https://doi.org/10.1158/0008-5472.CAN-10-2458.
Kumar A, Vigato C, LolliKumar DML. Phenothiazines as anti-cancer agents: SAR overview and synthetic strategies. Eur J Med Chem. 2023;254:115337. https://doi.org/10.1016/j.ejmech.2023.115337.
Nyarko RO, Awuchi CG, Kumar R, Boateng E, Kahwa I, Boateng PO, Saha P. Evaluation of cafeteria diet in experimental animal with plant extract of calotropis procera for obesity parameter. J for Res Appl Sci and Biotechnol. 2022;1(3):107–13.
Halder AK, Mallick S, Shikha D, Saha A, Krishna D, Jha T. Design of dual MMP-2/HDAC-8 inhibitors by pharmacophore map**, molecular docking, synthesis and biological activity. RSC Adv. 2015;5:72373–86.
Kumar S, Keshamma E, Trivedi U, Janjua D, Shaw P, Kumar R, Saha P. A meta analysis of different herbs (leaves, roots, stems) used in treatment of cancer cells. J for Res Appl Sci and Biotechnol. 2022;1(3):92–101.
De Cremoux P, Dalvai M, N’Doye O, Moutahir F, Rolland G, Chouchane-Mlik O, Assayag F, Lehmann-Che J, Kraus-Berthie L, Nicolas A, et al. HDAC inhibition does not induce estrogen receptor in human triple-negative breast cancer cell lines and patient-derived xenografts. Breast Cancer Res Treat. 2015;149:81–9. https://doi.org/10.1007/s10549-014-3233-y.
Dowling CM, Hollinshead KER, Di Grande A, Pritchard J, Zhang H, Dillon ET, Haley K, Papadopoulos E, Mehta AK, Bleach R, et al. Multiple screening approaches reveal HDAC6 as a novel regulator of glycolytic metabolism in triple-negative breast cancer. Sci Adv. 2021;7:eabc4897. https://doi.org/10.1126/sciadv.abc4897.
Min A, Im SA, Kim DK, Song SH, Kim HJ, Lee KH, Kim TY, Han SW, Oh DY, Kim TY, et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), enhances anti-tumor effects of the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib in triple-negative breast cancer cells. Breast Cancer Res. 2015;17:33. https://doi.org/10.1186/s13058-015-0534-y.
Sultana A, Singh M, Kumar A, Kumar R, Saha P, Kumar RS, Kumar D. To identify drug-drug interaction in cardiac patients in tertiary care hospitals. J Res Appl Sci and Biotechnol. 2022;1(3):146–52.
Bali P, Pranpat M, Swaby R, Fiskus W, Yamaguchi H, Balasis M, Rocha K, Wang HG, Richon V, Bhalla K. Activity of suberoylanilide hydroxamic acid against human breast cancer cells with amplification of her-2. Clin Cancer Res. 2005;11:6382–9. https://doi.org/10.1158/1078-0432.CCR-05-0344.
Gammoh N, Lam D, Puente C, Ganley I, Marks PA, Jiang X. Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc Natl Acad Sci USA. 2012;109:6561–5. https://doi.org/10.1073/pnas.1204429109.
Bellucci L, Dalvai M, Kocanova S, Moutahir F, Bystricky K. Activation of p21 by HDAC inhibitors requires acetylation of H2AZ. PLoS ONE. 2013;8:e54102. https://doi.org/10.1371/journal.pone.0054102.
Yang F, Zhang T, Wu H, Yang Y, Liu N, Chen A, Li Q, Li J, Qin L, Jiang B, et al. Design and optimization of novel hydroxamate-based histone deacetylase inhibitors of bis-substituted aromatic amides bearing potent activities against tumor growth and metastasis. J Med Chem. 2014;57:9357–69. https://doi.org/10.1021/jm5012148.
Yang F, Shan P, Zhao N, Ge D, Zhu K, Jiang CS, Li P, Zhang H. Developmenta of hydroxamate-based histone descetylase inhibitors containing 1,2,4-oxadiazole moiety core with antitumor activities. Bioorg Med Chem Lett. 2019;29:15–21. https://doi.org/10.1016/j.bmcl.2018.11.027.3.
Shan P, Yang F, Qi H, Hu Y, Zhu S, Sun Z, Zhang Z, Wang C, Hou C, Yu J, et al. Alteration of MDM2 by the small molecule YF438 exerts antitumor effects in triple-negative breast cancer. Cancer Res. 2021;81:4027–40. https://doi.org/10.1158/0008-5472.CAN-20-0922.
Yang F, Han L, Zhao N, Yang Y, Ge D, Zhang H, Chen Y. Synthesis and biological evaluation of thiophene-based hydroxamate derivatives as HDACis with antitumor activities. Future Med Chem. 2020;12:655–72. https://doi.org/10.4155/fmc-2019-0343.
Amle VS, Rathod DA, Keshamma E, Kumar V, Kumar R, Saha P. Bioactive herbal medicine use for eye sight: a meta analysis. J for Res Appl Sci and Biotechnol. 2022;1(3):42–50.
Yao D, Li C, Jiang J, Huang J, Wang J, He Z, Zhang J. Design, synthesis and biological evaluation of novel HDAC inhibitors with improved pharmacokinetic profile in breast cancer. Eur J Med Chem. 2020;205:112648. https://doi.org/10.1016/j.ejmech.2020.112648.
Sandhu P, Andrews P, Baker M, Koeplinger K, Soli E, Miller T, Baillie T. Disposition of vorinostat, a novel histone deacetylase inhibitor and anticancer agent, in preclinical species. Drug Metab Lett. 2007;1:153–61. https://doi.org/10.2174/187231207780363642.
Lee HY, Nepali K, Huang FI, Chang CY, Lai MJ, Li YH, Huang HL, Yang CR, Liou JP. (N-Hydroxycarbonylbenylamino)quinolines as selective histone deacetylase 6 inhibitors suppress growth of multiple myeloma in vitro and in vivo. J Med Chem. 2018;61:905–17. https://doi.org/10.1021/acs.jmedchem.7b01404.
Bresciani A, Ontoria JM, Biancofiore I, Cellucci A, Ciammaichella A, Di Marco A, Ferrigno F, Francone A, Malancona S, Monteagudo E, et al. Improved selective class I HDAC and novel selective HDAC3 inhibitors: beyond hydroxamic acids and benzamides. ACS Med Chem Lett. 2018;10:481–6. https://doi.org/10.1021/acsmedchemlett.8b00517.
Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–80.
Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–9.
Lee JH, Mahendran A, Yao Y, Ngo L, Venta-Perez G, Choy ML, Kim N, Ham WS, Breslow R, Marks PA. Development of a histone deacetylase 6 inhibitor and its biological effects. Proc Natl Acad Sci. 2013;110:15704–9.
Lee HY, Tsai AC, Chen MC, Shen PJ, Cheng YC, Kuo CC, Pan SL, Liu YM, Liu JF, Yeh TK, et al. Azaindolylsulfonamides, with a more selective inhibitory effect on histone deacetylase 6 activity, exhibit antitumor activity in colorectal cancer HCT116 cells. J Med Chem. 2014;57:4009–22.
Lee SH, Yoo C, Im S, Jung JH, Choi HJ, Yoo J. Expression of histone deacetylases in diffuse large B-cell lymphoma and its clinical significance. Int J Med Sci. 2014;11:994–1000.
Li Z, Zhu WG. Targeting histone deacetylases for cancer therapy: from molecular mechanisms to clinical implications. Int J Biol Sci. 2014;10:757–70.
Kumar R, Saha P, Keshamma E, Sachitanadam P, Subramanian M. Docking studies of some novel hetrocyclic compound as Acat inhibitors: a meta analysis. J for Res Applied Sci Biotechnol. 2022;1(3):33–41.
Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, Phelan C, Lazar MA. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 2006;20:2566–79.
Li D, Sun X, Zhang L, Yan B, **e S, Liu R, Liu M, Zhou J. Histone deacetylase 6 and cytoplasmic linker protein 170 function together to regulate the motility of pancreatic cancer cells. Protein Cell. 2014;5:214–23.
Li S, Liu X, Chen X, Zhang L, Wang X. Histone deacetylase 6 promotes growth of glioblastoma through inhibition of SMAD2 signaling. Tumour Biol. 2015;36:9661–5.
Li Y, Peng L, Seto E. HDAC10 regulates cell cycle G2/M phase transition via a novel Let-7-HMGA2-Cyclin A2 pathway. Mol Cell Biol. 2015;35:3547–65.
Liao W, Jordaan G, Srivastava MK, Dubinett S, Sharma S, Sharma S. Effect of epigenetic histone modifications on E-cadherin splicing and expression in lung cancer. Am J Cancer Res. 2013;3:374–89.
Liffers K, Kolbe K, Westphal M, Lamszus K, Schulte A. Histone deacetylase inhibitors resensitize EGFR/EGFRvIII-overexpressing, Erlotinib-resistant glioblastoma cells to tyrosine kinase inhibition. Target Oncol. 2015;11:29–40.
Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1α. Mol Cell. 2010;38:864–78.
Lin YH, Yuan J, Pei H, Liu T, Ann DK, Lou Z. KAP1 Deacetylation by SIRT1 promotes non-homologous end-joining repair. PLoS ONE. 2015;10: e0123935.
Liu KP, Zhou D, Ouyang DY, Xu LH, Wang Y, Wang LX, Pan H, He XH. LC3B-II deacetylation by histone deacetylase 6 is involved in serum-starvation-induced autophagic degradation. Biochem Biophys Res Commun. 2013;441:970–5.
Lobera M, Madauss KP, Pohlhaus DT, Wright QG, Trocha M, Schmidt DR, Baloglu E, Trump RP, Head MS, Hofmann GA, et al. Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat Chem Biol. 2013;9:319–25.
Locatelli SL, Cleris L, Stirparo GG, Tartari S, Saba E, Pierdominici M, Malorni W, Carbone A, Anichini A, Carlo-Stella C. BIM upregulation and ROS-dependent necroptosis mediate the antitumor effects of the HDACi Givinostat and Sorafenib in Hodgkin lymphoma cell line xenografts. Leukemia. 2014;28:1861–71.
Lopez G, Bill KL, Bid HK, Braggio D, Constantino D, Prudner B, Zewdu A, Batte K, Lev D, Pollock RE. HDAC8, A potential therapeutic target for the treatment of malignant peripheral nerve sheath tumors (MPNST). PLoS ONE. 2015;10: e0133302.
Luna A, Aladjem MI, Kohn KW. SIRT1/PARP1 crosstalk: Connecting DNA damage and metabolism. Genome Integr. 2013;4:6.
Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–48.
Lv Z, Weng X, Du C, Zhang C, **ao H, Cai X, Ye S, Cheng J, Ding C, **e H, et al. Downregulation of HDAC6 promotes angiogenesis in hepatocellular carcinoma cells and predicts poor prognosis in liver transplantation patients. Mol Carcinog. 2015;55:1024–33.
Mai A, Massa S, Pezzi R, Simeoni S, Rotili D, Nebbioso A, Scognamiglio A, Altucci L, Loidl P, Brosch G. Class II (IIa)-selective histone deacetylase inhibitors. 1: synthesis and biological evaluation of novel (aryloxopropenyl)pyrrolyl hydroxyamides. J Med Chem. 2005;48:3344–53.
Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc Natl Acad Sci. 2013;110:2647–52.
Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–52.
Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–6.
Marek L, Hamacher A, Hansen FK, Kuna K, Gohlke H, Kassack MU, Kurz T. Histone deacetylase (HDAC) inhibitors with a novel connecting unit linker region reveal a selectivity profile for HDAC4 and HDAC5 with improved activity against chemoresistant cancer cells. J Med Chem. 2013;56:427–36.
Marquard L, Poulsen CB, Gjerdrum LM, de Nully BP, Christensen IJ, Jensen PB, Sehested M, Johansen P, Ralfkiaer E. Histone deacetylase 1, 2, 6 and acetylated histone H4 in B- and T-cell lymphomas. Histopathology. 2009;54:688–98.
Matsuba S, Niwa S, Muraki K, Kanatsuka S, Nakazono Y, Hatano N, Fujii M, Zhan P, Suzuki T, Ohya S. Downregulation of Ca2+-activated Cl- channel TMEM16A by the inhibition of histone deacetylase in TMEM16A-expressing cancer cells. J Pharmacol Exp Ther. 2014;351:510–8.
Marquard L, Gjerdrum LM, Christensen IJ, Jensen PB, Sehested M, Ralfkiaer E. Prognostic significance of the therapeutic targets histone deacetylase 1, 2, 6 and acetylated histone H4 in cutaneous T-cell lymphoma. Histopathology. 2008;53:267–77.
McDermott J, Jimeno A. Belinostat for the treatment of peripheral T-cell lymphomas. Drugs Today (Barc). 2014;50:337–45.
McGraw AL. Romidepsin for the treatment of T-cell lymphomas. Am J Health Syst Pharm. 2013;70:1115–22.
Meidhof S, Brabletz S, Lehmann W, Preca BT, Mock K, Ruh M, Schuler J, Berthold M, Weber A, Burk U, et al. ZEB1-associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol Med. 2015;7:831–47.
Methot JL, Chakravarty PK, Chenard M, Close J, Cruz JC, Dahlberg WK, Fleming J, Hamblett CL, Hamill JE, Harrington P, et al. Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2). Bioorg Med Chem Lett. 2008;18:973–8.
Milde T, Oehme I, Korshunov A, Kopp-Schneider A, Remke M, Northcott P, Deubzer HE, Lodrini M, Taylor MD, von Deimling A, et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16:3240–52.
Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–51.
Min SK, Koh YH, Park Y, Kim HJ, Seo J, Park HR, Cho SJ, Kim IS. Expression of HAT1 and HDAC1, 2, 3 in diffuse large B-cell lymphomas, peripheral T-cell lymphomas, and NK/T-cell lymphomas. Korean J Pathol. 2012;46:42–150.
Minami J, Suzuki R, Mazitschek R, Gorgun G, Ghosh B, Cirstea D, Hu Y, Mimura N, Ohguchi H, Cottini F, et al. Histone deacetylase 3 as a novel therapeutic target in multiple myeloma. Leukemia. 2014;28:680–9.
Cheng Y, He C, Wang M, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Sig Transduct Target Ther. 2019;4:62. https://doi.org/10.1038/s41392-019-0095-0.
Minamiya Y, Ono T, Saito H, Takahashi N, Ito M, Motoyama S, Ogawa J. Strong expression of HDAC3 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Tumour Biol. 2010;31:533–9.
Minamiya Y, Ono T, Saito H, Takahashi N, Ito M, Mitsui M, Motoyama S, Ogawa J. Expression of histone deacetylase 1 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Lung Cancer. 2011;74:300–4.
Mishima Y, Santo L, Eda H, Cirstea D, Nemani N, Yee AJ, O’Donnell E, Selig MK, Quayle SN, Arastu-Kapur S, et al. Ricolinostat (ACY-1215) induced inhibition of aggresome formation accelerates carfilzomib-induced multiple myeloma cell death. Br J Haematol. 2015;169:423–34.
Nyarko RO, Awuchi CG, Kumar R, Boateng E, Kahwa I, Boateng PO, Saha P. Evaluation of cafeteria diet in experimental animal with plant extract of calotropis procera for obesity parameter. J Res Appl Sci and Biotechnol. 2022;1(3):107–13.
Mithraprabhu S, Kalff A, Chow A, Khong T, Spencer A. Dysregulated class I histone deacetylases are indicators of poor prognosis in multiple myeloma. Epigenetics. 2014;9:1511–20.
Subramanian M, Keshamma E, Janjua D, Kumar D, Kumar R, Saha P, Rao S. Quality risk management approach for drug development and its future prospectives. J for Res Appl Sci Biotechnol. 2022;1(3):166–77.
Wapenaar H, Dekker FJ. Histone acetyltransferases: challenges in targeting bi-substrate enzymes. Clin Epigenet. 2016;8:59.
Moffat D, Patel S, Day F, Belfield A, Donald A, Rowlands M, Wibawa J, Brotherton D, Stimson L, Clark V, et al. Discovery of 2-(6-{[(6-fluoroquinolin-2-yl)methyl]amino}bicyclo[3.1.0]hex-3-yl)-N-hydroxypyrimidine-5-carboxamide (CHR-3996), a class I selective orally active histone deacetylase inhibitor. J Med Chem. ;53:8663–8678.
Moreno DA, Scrideli CA, Cortez MA, de Paula QR, Valera ET, da Silva SV, Yunes JA, Brandalise SR, Tone LG. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br J Haematol. 2010;150:665–73.
Moresi V, Carrer M, Grueter CE, Rifki OF, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. Proc Natl Acad Sci. 2012;109:1649–54.
Morschhauser F, Terriou L, Coiffier B, Bachy E, Varga A, Kloos I, Lelievre H, Sarry AL, Depil S, Ribrag V. Phase 1 study of the oral histone deacetylase inhibitor abexinostat in patients with Hodgkin lymphoma, non-Hodgkin lymphoma, or chronic lymphocytic leukaemia. Invest New Drugs. 2015;33:423–31.
Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–29.
Muller BM, Jana L, Kasajima A, Lehmann A, Prinzler J, Budczies J, Winzer KJ, Dietel M, Weichert W, Denkert C. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer–overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer. 2013;13:215.
Munster PN, Thurn KT, Thomas S, Raha P, Lacevic M, Miller A, Melisko M, Ismail-Khan R, Rugo H, Moasser M, et al. A phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy-resistant breast cancer. Br J Cancer. 2011;104:1828–35.
Li Y, Seto E. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb Perspect Med. 2016;6(10):a026831. https://doi.org/10.1101/cshperspect.a026831.
Kawai H, Li H, Avraham S, Jiang S, Avraham HK. Overexpression of histone deacetylase HDAC1 modulates breast cancer progression by negative regulation of estrogen receptor alpha. Int J Cancer. 2003;107:353–8.
Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Hara Y, Kobayashi S, Iwase H. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast*. Breast Cancer Res Treat. 2005;94:11–6.
Khochbin S, Verdel A, Lemercier C, Seigneurin-Berny D. Functional significance of histone deacetylase diversity. Curr Opin Genet Dev. 2001;11:162–6.
Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL, Eckert KA, Verderame MF, Welch DR. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the MSIN3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279:1562–9.
Garmpi A, Garmpis N, Damaskos C, Valsami S, Spartalis E, Lavaris A, Patelis N, Margonis GA, Apostolou KG, Spartalis M, et al. Histone deacetylase inhibitors as a new anticancer option: How far can we go with expectations? J BUON. 2018;23:846–61.
Damaskos C, Karatzas T, Nikolidakis L, Kostakis ID, Karamaroudis S, Boutsikos G, Damaskou Z, Kostakis A, Kouraklis G. Histone deacetylase (HDAC) inhibitors: current evidence for therapeutic activities in pancreatic cancer. Anticancer Res. 2015;35:3129–35.
Marks PA. The clinical development of histone deacetylase inhibitors as targeted anticancer drugs. Expert Opin Investig Drugs. 2010;19:1049–66.
Ruefli AA, Ausserlechner MJ, Bernhard D, Sutton VR, Tainton KM, Kofler R, Smyth MJ, Johnstone RW. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proc Natl Acad Sci USA. 2001;98:10833–8.
Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97:10014–9.
Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–9.
Qian DZ, Kachhap SK, Collis SJ, Verheul HM, Carducci MA, Atadja P, Pili R. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1. Cancer Res. 2006;66:8814–21.
Kano Y, Akutsu M, Tsunoda S, Izumi T, Kobayashi H, Mano H, Furukawa Y. Cytotoxic effects of histone deacetylase inhibitor FK228 (depsipeptide, formally named FR901228) in combination with conventional anti-leukemia/lymphoma agents against human leukemia/lymphoma cell lines. Investig New Drugs. 2006;25:31–40.
Wardell SE, Ilkayeva OR, Wieman HL, Frigo DE, Rathmell JC, Newgard CB, McDonnell DP. Glucose metabolism as a target of histone deacetylase inhibitors. Mol Endocrinol. 2009;23:388–401.
Barbarotta L, Hurley K. Romidepsin for the treatment of peripheral T-cell lymphoma. J Adv Pract Oncol. 2015;6:22–36.
Libby EN, Becker PS, Burwick N, Green DJ, Holmberg L, Bensinger WI. Panobinostat: a review of trial results and future prospects in multiple myeloma. Expert Rev Hematol. 2014;8:9–18.
Damaskos C, Garmpis N, Valsami S, Kontos M, Spartalis E, Kalampokas T, Kalampokas E, Athanasiou A, Moris D, Daskalopoulou A, et al. Histone deacetylase inhibitors: an attractive therapeutic strategy against breast cancer. Anticancer Res. 2017;37:35–46.
Woo YM. Epigenetic regulation in cystogenesis. Adv Exp Med Biology. 2016;933:59–68.
Mann BS, Johnson JR, He K, Sridhara R, Abraham S, Booth BP, Verbois L, Morse DE, Jee JM, Pope S, Harapanhalli RS, Dagher R, Farrell A, Justice R, Pazdur R. Vorinostat for treatment of cutaneous manifestations of advanced primary cutaneous T-cell lymphoma. Clin Cancer Res. 2007;13:2318–22.
Marks PA. Discovery and development of SAHA as an anticancer agent. Oncogene. 2007;26:1351–6.
Damaskos C, Valsami S, Spartalis E, Antoniou EA, Tomos P, Karamaroudis S, Zoumpou T, Pergialiotis V, Stergios K, Michaelides C, Kontzoglou K, Perrea D, Nikiteas N, Dimitroulis D. Histone deacetylase inhibitors: a novel therapeutic weapon against medullary thyroid cancer? Anticancer Res. 2016;36:5019–24.
Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays. 1995;17:423–30.
Blumenschein GR Jr, Kies MS, Papadimitrakopoulou VA, Lu C, Kumar AJ, Ricker JL, Chiao JH, Chen C, Frankel SR. Phase II trial of the histone deacetylase inhibitor vorinostat (Zolinza, suberoylanilide hydroxamic acid, SAHA) in patients with recurrent and/or metastatic head and neck cancer. Invest New Drugs. 2008;26:81–7.
Luu TH, Morgan RJ, Leong L, Lim D, McNamara M, Portnow J, Frankel P, Smith DD, Doroshow JH, Wong C, Aparicio A, Gandara DR, Somlo G. A phase II trial of vorinostat (suberoylanilide hydroxamic acid) in metastatic breast cancer: a California Cancer Consortium study. Clin Cancer Res. 2008;14:7138–42.
Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–93.
Munster PN, Troso-Sandoval T, Rosen N, Rifkind R, Marks PA, Richon VM. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces differentiation of human breast cancer cells. Cancer Res. 2001;61:8492–7.
Bali P, Pranpat M, Swaby R, Fiskus W, Yamaguchi H, Balasis M, Rocha K, Wang HG, Richon V, Bhalla K. Activity of suberoylanilide hydroxamic acid against human breast cancer cells with amplification of HER2. Clin Cancer Res. 2005;11:6382–9.
Wu S, Luo Z, Yu PJ, **e H, He YW. Suberoylanilide hydroxamic acid (SAHA) promotes the epithelial–mesenchymal transition of triple-negative breast cancer cells via HDAC8/FOXA1 signals. Biol Chem. 2016;397:75–83.
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial– mesenchymal transitions in development and disease. Cell. 2009;139:871–90.
Sami S, Hoti N, Xu HM, Shen Z, Huang X. Valproic acid inhibits the growth of cervical cancer both in vitro and in vivo. J Biochem. 2008;144:357–62.
Kurwale N, Garg K, Arora A, Chandra PS, Tripathi M. Valproic acid as an antiepileptic drug: Is there a clinical relevance for the epilepsy surgeon? Epilepsy Res. 2016;127:191–4.
Peselow ED, Clevenger S, IsHak WW. Prophylactic efficacy of lithium, valproic acid, and carbamazepine in the maintenance phase of bipolar disorder: a naturalistic study. Int Clin Psychopharmacol. 2016;31:218–23.
Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S, Sleeman JP, Lo Coco F, Nervi C, Pelicci PG, Heinzel T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–78.
Hrzenjak A, Moinfar F, Kremser ML, Strohmeier B, Staber PB, Zatloukal K, Denk H. Valproate inhibition of histone deacetylase 2 affects differentiation and decreases proliferation of endometrial stromal sarcoma cells. Mol Cancer Ther. 2006;5:2203–10.
Fortunati N, Bertino S, Costantino L, Bosco O, Vercellinatto I, Catalano MG, Boccuzzi G. Valproic acid is a selective antiproliferative agent in estrogen-sensitive breast cancer cells. Cancer Lett. 2008;259:156–64.
Travaglini L, Vian L, Billi M, Grignani F, Nervi C. Epigenetic reprogramming of breast cancer cells by valproic acid occurs regardless of estrogen receptor status. Int J Biochem Cell Biol. 2009;41:225–34.
Zhang L, Wang G, Wang L, Song C, Leng Y, Wang X, Kang J. VPA inhibits breast cancer cell migration by specifically targeting HDAC2 and down-regulating Survivin. Mol Cell Biochem. 2012;361:39–45.
Mawatari T, Ninomiya I, Inokuchi M, Harada S, Hayashi H, Oyama K, Makino I, Nakagawara H, Miyashita T, Tajima H, Takamura H, Fushida S, Ohta T. Valproic acid inhibits proliferation of HER2-expressing breast cancer cells by inducing cell cycle arrest and apoptosis through HSP70 acetylation. Int J Oncol. 2015;47:2073–81.
Ganai SA. Panobinostat: The small molecule metalloenzyme inhibitor with marvelous anticancer activity. Curr Top Med Chem. 2016;16:427–34.
Zhou Q, Atadja P, Davidson NE. Histone deacetylase inhibitor LBH589 reactivates silenced estrogen receptor alpha (ER) gene expression without loss of DNA hypermethylation. Cancer Biol Ther. 2007;6:64–9.
Prestegui-Martel B, Bermudez-Lugo JA, Chavez-Blanco A, Duenas-Gonzalez A, Garcia-Sanchez JR, Perez-Gonzalez OA, Padilla-Martinez II, Fragoso-Vazquez MJ, Mendieta-Wejebe JE, Correa-Basurto AM, Mendez-Luna D, Trujillo-Ferrara J, Correa-Basurto J. N-(2-Hydroxyphenyl)-2-propylpentanamide, a valproic acid aryl derivative designed in silico with improved anti-proliferative activity in HeLa, rhabdomyosarcoma and breast cancer cells. J Enzyme Inhib Med Chem. 2016;2:1–10.
Arun K, Chiara V, Donatella B, Marco LL, Deepak K. Phenothiazines as anti-cancer agents: SAR overview and synthetic strategies. Eur J Med Chem. 2023;254: 115337.
Kumar A, Yadav AK, Mishra V, Kumar D. Recent Advancements in Triazole-based Click Chemistry in Cancer Drug Discovery and Development. SynOpen. 2023;7:186–208.
The National Institute of Health. Clinical Trials database; 2019. https://clinicaltrials.gov/ct2/home
Wong KK, Fracasso PM, Bukowski RM, Lynch TJ, Munster PN, Shapiro GI, Jänne PA, Eder JP, Naughton MJ, Ellis MJ, Jones SF. A phase I study with neratinib (HKI-272), an irreversible pan ErbB receptor tyrosine kinase inhibitor, in patients with solid tumors. Clin Cancer Res. 2009;15(7):2552–8.
King SE, Skinner MK. Epigenetic transgenerational inheritance of obesity susceptibility. Trends Endocrinol Metab. 2020;31(7):478–94.
Wang JY, Minami Y, Zhu J. Abl and Cell Death. Abl Family Kinases in Development and Disease. 2006;26–47.
Jasek K, Kasubova I, Holubekova V, Stanclova A, Plank L, Lasabova Z. Epigenetics: an alternative pathway in GISTs tumorigenesis. Neoplasma. 2018;65(4):477–93.
Luo G, Hu Y, Zhang Z, Wang P, Luo Z, Lin J, Cheng C, Yang Y. Clinicopathologic significance and prognostic value of Ki-67 expression in patients with gastric cancer: a meta-analysis. Oncotarget. 2017;8(30):50273.
Weiss AJ, Iqbal J, Zaidi N, Mechanick JI. The skeletal subsystem as an integrative physiology paradigm. Curr Osteoporos Rep. 2010;8:168–77.
Burley SK, Berman HM, Kleywegt GJ, Markley JL, Nakamura H, Velankar S. 2017 Protein Data Bank (PDB): the single global macromolecular structure archive. Protein crystallography: methods and protocols. 627–41.
Micelli C, Rastelli G. Histone deacetylases: structural determinants of inhibitor selectivity. Drug Discovery Today. 2015;20(6):718–35.
Morris GM, Huey R, Olson AJ. Using autodock for ligand receptor docking. Curr Protoc Bioinformatics. 2008;24(1):8–14.
Pymol DeLano WL. An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002;40(1):82–92.
Jejurikar BL, Rohane SH. Drug designing in discovery studio.
Raghuvanshi D, Kumar S, Shukla MK, Kumar D, Kumar D, Verma R, Nepovimova E, Valko M, Alomar SY, Alwasel SH, Kuca K. Assessment of phytochemicals, antioxidants and in-silico molecular dynamic simulation of plant derived potential inhibitory activity of Thalictrum. Biomed. Pharmacother. 2022;156:113898–917. https://doi.org/10.1016/j.biopha.2022.113898.
Kumar S, Sengupta S, Ali I, Gupta MK, Lalhlenmawia H, Azizov S, Kumar D. Identification and exploration of quinazoline-1,2,3-triazole inhibitors targeting EGFR in lung cancer. J Biomol Struct Dyn. 2023. https://doi.org/10.1080/07391102.2023.2204360.
Vimal SK, Cao H, Dubey A, Agrawal L, Pathak N, Zuo H, Kumar D, Bhattacharyya S. In vivo and in silico investigations of the pegylated gold nanoparticle treatment of amyotrophic lateral sclerosis in mice. New J Chem. 2022;46(25):12252–64.
Huey R, Morris GM, Forli S. Using AutoDock 4 and Vina with AutoDockTools: A Tutorial. Scripps Research Institute, California, USA. 2011 Dec 8.
Pawar RP, Rohane SH. Role of autodock vina in PyRx molecular docking. Asian J. Research Chem. 2021;14(2):132–4.
Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK. 2006 Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. 84
Madhavi Sastry G, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–34.
Steinmann SN, Sautet P, Michel C. Solvation free energies for periodic surfaces: comparison of implicit and explicit solvation models. Phys Chem Chem Phys. 2016;18(46):31850–61.
Acknowledgements
The authors are thankful to their respective institutes for the support.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that there is no conflict of interest.
Ethical approval
Not applicable.
Informed consent
We agreed with the journal policy and provided our consent for the publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mehmood, S.A., Sahu, K.K., Sengupta, S. et al. Recent advancement of HDAC inhibitors against breast cancer. Med Oncol 40, 201 (2023). https://doi.org/10.1007/s12032-023-02058-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-023-02058-x