Abstract
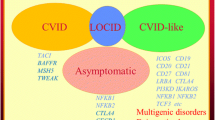
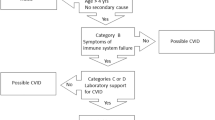
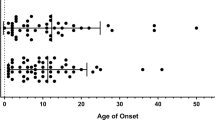
Identification of the causes of monogenetic common variable immunodeficiency (CVID) patients has rapidly increased in the last years by means of worldwide availability of appropriate genetic diagnostic methods. However, up to date, very limited numbers of reports demonstrating the role of geography, ethnicity, and consanguinity have been published. Here, we reported the first study of Turkish CVID patients and compared them with the results of three countries from America, Europe, and Asia. A total of 100 children diagnosed as CVID according to the criteria of European Society for Immunodeficiencies were enrolled, and they were genetically analyzed by using targeted next-generation sequencing and whole-exome sequencing. The median age of our patients was 5.8 years (range, 3.0–16.0 years) at clinical diagnosis and 9.0 years (range, 4.8–21.0 years) at the time of genetic diagnosis. The consanguinity rate was 24%. Disease-causing pathogenic variants were defined in 40% of patients in a total of 17 different genes. Sixteen of 40 identified pathogenic variants were novel (40%). We determined 18 surface molecular defects, 10 cytosolic defects, 9 nuclear defects, and 3 others. In our cohort, the most common gene was TACI (15/40 in pathogenic variant identified cases and 15/100 in all cases) followed by the others such as PLCү2, LRBA, TCF3, and STAT1. In contrast to our expectations, our results were more similar to American and European population rather than Asians, although we also have high consanguinity rates and live on the geography between Europe and Asia. Genetic investigation is a great challenge, because of the complexity and heterogeneity of the disease, and each country has to know their own current genetic landscape in CVID for a better and successful management of the patients.

Similar content being viewed by others
Data availability
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Abbreviations
- CVID:
-
Common variable immunodeficiency
- ACMG:
-
American College of Medical Genetics and Genomics
- VUS:
-
Variant of unknown significance
- PM2:
-
Moderate evidence of pathogenicity absent from controls (or at extremely low frequency if recessive) in Exome Sequencing Project, 1000 Genomes, or ExAC
- PM1:
-
Moderate evidence of pathogenicity located in a mutational hot spot and/or critical and well-established functional domain (e.g. active site of an enzyme) without benign variation
- PP2:
-
Missense variant in a gene that has a low rate of benign missense variation and where missense variants are a common mechanism of disease
- PP3:
-
Supporting evidence of pathogenicity where multiple lines of computational evidence support a deleterious effect on the gene or gene product (conservation, evolutionary, splicing impact, etc.)
- BP4:
-
Multiple lines of computational evidence suggest no impact on gene or gene product (conservation, evolutionary, splicing impact, etc.)
- TNGS:
-
Targeted next-generation sequencing
- WES:
-
Whole-exome sequencing
References
Bogaert DJ, Dullaers M, Lambrecht BN, Vermaelen KY, De Baere E, Haerynck F. Genes associated with common variable immunodeficiency: one diagnosis to rule them all. J Med Genet. 2016;53:575–90.
Li J, Wei Z, Li YR, Maggadottir SM, Chang X, Desai A, Hakonarson H. Understanding the genetic and epigenetic basis of common variable immunodeficiency disorder through omics approaches. Biochim Biophys Acta. 2016;1860:2656–63.
Li R, Zheng Y, Li Y, Zhang R, Wang F, Yang D, Ma Y, Mu X, Cao Z, Gao Z. Common variable immunodeficiency with genetic defects identified by whole exome sequencing. Biomed Res Int. 2018;2018:3724630. https://doi.org/10.1155/2018/3724630.
Yazdani R, Habibi S, Sharifi L, Azizi G, Abolhassani H, Olbrich P, Aghamohammadi A. Common variable immunodeficiency: epidemiology, pathogenesis, clinical manifestations, diagnosis, classification and management. J Investig Allergol Clin Immunol. 2020;30:14–34.
Abolhassani H, Hammarstrom L, Cunningha-Rundles C. Current genetic landscape in common variable immune deficiency. Blood. 2020;135:656–67.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010;6: e1001025.
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;7:20–30.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81.
Karaca NE, Severcan EU, Guven B, Azarsiz E, Aksu G, Kutukculer N. TNFRSF13B/TACI alterations in turkish patients with common variable immunodeficiency and IgA deficiency. Avicenna J Med Biotechnol. 2018;10:192–5.
Edeer Karaca N, Özek G, Ataseven E, Tökmeci N, Şenol HD, Kıran E. Combined immunodeficiency with marginal zone lymphoma due to a novel homozygous mutation in IL-21R gene and successful treatment with hematopoietic stem cell transplantation. Pediatr Hematol Oncol. 2021;38:745–52.
Kutukculer N, Topyildiz E, Berdeli A, Guven Bilgin B, Aykut A, Durmaz A, Cogulu O, et al. Four diseases, PLAID, APLAID, FCAS3 and CVID and one gene (PHOSPHOLIPASE C, GAMMA-2; PLCG2): striking clinical phenotypic overlap and difference. Clin Case Rep. 2021;9:2023–31.
Eren Akarcan S, Edeer Karaca N, Aksu G, Aykut A, Yilmaz Karapinar D, Cetin F, et al. Two male siblings with a novel LRBA mutation presenting with different findings of IPEX syndrome. JMM Case Rep. 2018;5:e005167. https://doi.org/10.1099/jmmcr.0.005167.
Jamee M, Azizi G, Baris S, Karakoc-Aydiner E, Ozen A, Kiliç SŞ, et al. Clinical, immunological, molecular and therapeutic findings in monogenic immune dysregulation diseases: Middle East and North Africa registry. J Clin Immunol. 2022;244:109131. https://doi.org/10.1016/j.clim.2022.109131.
Eren Akarcan S, Ulusoy Severcan E, Edeer Karaca N, Isik E, Aksu G, Migaud M, et al. Gain-of-function mutations in STAT1: a recently defined cause for chronic mucocutaneous candidiasis disease mimicking combined immunodeficiencies. Case Rep in Immunol. 2017;2846928. https://doi.org/10.1155/2017/2846928.
Edeer Karaca N, Aykut A, Parıltay E, Aksu G, Cogulu Ö, Kutukculer N. A novel TTC37 mutation causing clinical symptoms of trichohepatoenteric syndrome such as pyoderma gangrenosum and immunodeficiency without severe diarrhea. J Investig Allergol Clin Immunol. 2019;29:396–8.
Driessen GJ, IJspeert H, Wentink M, Yntema HG, van Hagen PM, van Strien A, et al. Increased PI3K/Akt activity and deregulated humoral immune response in human PTEN deficiency. J Allergy Clin Immunol. 2016;138:1744–7.
Topyıldız E, Karaca NE, Bas I, Aykut A, Durmaz A, Bilgin RBG, Aksu G, Karapınar DY, Kutukculer N. A novel homozygous TRNT1 mutation in a child with an early diagnosis of common variable immunodeficiency leading to mild hypogammaglobulinemia and hemolytic anemia. J Pediatr Hematol Oncol. 2021;43:e780–4.
Kutukculer N, Seeholzer T, O’Neill TJ, Graß C, Aykut A, Karaca NE, Durmaz A, Cogulu O, Aksu G, Gehring T, Gewies A, Krappmann D. Human immune disorder associated with homozygous hypomorphic mutation affecting MALT1B splice variant. J Allergy Clin Immunol. 2021;147:775–8.
Kutukculer N, Gulez N, Karaca NE, Aksu G, Berdeli A. Three different classifications, B lymphocyte subpopulations, TNFRSF13B (TACI), TNFRSF13C (BAFF-R), TNFRSF13 (APRIL) gene mutations, CTLA-4 and ICOS gene polymorphisms, in Turkish patients with common variable immunodeficiency. J Clin Immunol. 2012;32:1165–79.
Karaca NE, Severcan EU, Bilgin BG, Azarsiz E, Akarcan S, Gunaydın NC, et al. Familial inheritance and screening of first-degree relatives in common variable immunodeficiency and immunoglobulin A deficiency patients. Int J Immunopathol Pharmacol. 2018;32:2058738418779458. https://doi.org/10.1177/2058738418779458.
Bisgin A, Sonmezler O, Boga I, Yilmaz M. The impact of rare and low-frequency genetic variants in common variable immunodeficiency (CVID). Sci Rep. 2021;11:8308. https://doi.org/10.1038/s41598-021-87898-1.
Shi C, Wang F, Tong A, Zhang XQ, Song HM, Liu ZY, Lyu W, Liu YH, **a WB. NFKB2 mutation in common variable immunodeficiency and isolated adrenocorticotropic hormone deficiency: a case report and review of literature. Medicine (Baltimore). 2016;95(40):e5081. https://doi.org/10.1097/MD.0000000000005081.
Luo MZ, Xu T, Hue XH, Wang YP, Wu PL, Chen XM, Tang XM, Zao XD, Zhang ZY. De novo NFKB2 gene mutation associated with common variable immunodeficiency. Zhonghua Er Ke Za Zhi. 2018;56:628–32.
Lee JJ, Özcan E, Rauter I, Geha RS. Transmembrane activator and calcium-modulator and cyclophilin ligand interactor mutations in common variable immunodeficiency. Curr Opin Allergy Clin Immunol. 2008;8:520–6.
Kaskas I, Tsinti G, Kalala F, Farmaki E, Kourakli A, Kapousouzi A, et al. TACI mutations in antibody deficiencies. A nationwide study in Greece. Medicina. 2021;57:827–35.
Firtina S, Ng YY, Ng OH, Kiykim A, Ozek EY, Kara M, et al. Primary antibody deficiencies in Turkey: molecular and clinical aspects. Immunol Res. 2022;70:44–55.
Maffucci P, Filion CA, Boisson B, Itan Y, Shang L, Casanova JL, et al. Genetic diagnosis using whole exome sequencing in common variable immunodeficiency. Front Immunol. 2016;7:220–6.
Kedar P, Dongerdiye R, Chandrakala S, Bargir UA, Madkaikar M. Targeted next-generation sequencing revealed a novel homozygous mutation in the LRBA gene causes severe hemolysis associated with inborn errors of immunity. Hematology. 2022;27:441–8.
Acknowledgements
The authors thank the Jeffrey Modell Foundation for their precious support.
Funding
This work was partly supported by the Jeffrey Modell Foundation (JMF).
Author information
Authors and Affiliations
Contributions
Ayse Aygun: study conception, design, and write the manuscript. Ezgi Topyıldız: data collection. Necil Kutukculer, Neslihan Edeer Karaca: analysis and interpretation of results, data collection. Mehmet Geyik: material preparation. Asude Durmaz, Guzide Aksu, Ayca Aykut: data collection. Necil Kutukculer: study design, supervised the work, performed the analysis, contributed data, and analysis tools. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
The study was approved by the local ethics committee (Ege University Faculty of Medicine Clinical Ethical Board Approval No: 23-7.IT/33).
Consent to participate
Written informed consent was obtained from the parents or legal guardians.
Consent for publication
The authors affirm that parents or legal guardians provided informed consent for publication.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Aygun, A., Topyıldız, E., Geyik, M. et al. Current genetic defects in common variable immunodeficiency patients on the geography between Europe and Asia: a single-center experience. Immunol Res 72, 225–233 (2024). https://doi.org/10.1007/s12026-023-09426-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-023-09426-9