Abstract
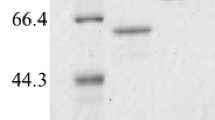
A novel amylase AmyFlA from Flavobacterium sp. NAU1659, AmyFlA, was cloned and expressed in Esherichia coli. Based on phylogenetic and functional analysis, it was identified as a novel member of the subfamily GH13_46, sharing high sequence identity. The protein was predicted to consist of 620 amino acids, with a putative signal peptide of 25 amino acids. The enzyme was able to hydrolyze soluble starch with a specific activity of 352.97 U/mg at 50 °C in 50 mM phosphate buffer (pH 6.0). The Km and Vmax values of AmyFlA were respectively 3.15 mg/ml and 566.36 µmol·ml−1·min−1 under optimal conditions. Its activity towards starch was enhanced by 63% in the presence of 1 mM Ca2+, indicating that AmyFlA was a Ca2+-dependent amylase. Compared to the reported maltogenic amylases, AmyFlA produced a lower variety of intermediate oligosaccharides at the start of the reaction so that the product mixture contained a higher proportion of maltose. These results indicate that AmyFlA may be potential application value in the production of high-maltose syrup.






Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this paper.
References
Wang, S., Li, C., Copeland, L., Niu, Q., & Wang, S. (2015). Starch retrogradation: A comprehensive review. Comprehensive Reviews in Food Science Food Safety, 14, 568–585.
Sjöö, M., & Nilsson, L. (2017). Starch in food: Structure, function and applications. ed. Woodhead Publishing.
Pan, S., Ding, N., Ren, J., Gu, Z., Li, C., Hong, Y., Cheng, L., Holler, T. P., & Li, Z. (2017). Maltooligosaccharide-forming amylase: Characteristics, preparation, and application. Biotechnology Advances, 35, 619–632.
Smits, A., Kruiskamp, P., Van Soest, J., & Vliegenthart, J. (2003). The influence of various small plasticisers and malto-oligosaccharides on the retrogradation of (partly) gelatinised starch. Carbohydrate Polymers, 51, 417–424.
Chegeni, M., & Hamaker, B. (2015). Induction of differentiation of small intestinal enterocyte cells by maltooligosaccharides. The FASEB Journal, 29, 596514.
Ielux, J., Franco, O., Silva, M., Silvinski, T., Bloch Jr, C., Rigden, J., & de Sa, M. G. (2000). Purification, biochemical characterization and partial primary structure of a new a-amylase inhibitor from Secale cereale (rye). The International Journal of Biochemistry & Cell Biology, 32, 1195–1204.
Fulton, D. C., Stettler, M., Mettler, T., Vaughan, C. K., Li, J., Francisco, P., Gil, M., Reinhold, H., Eicke, S., & Messerli, G. (2008). β-Amylase4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. The Plant Cell, 20, 1040–1058.
Kumar, P., & Satyanarayana, T. (2009). Microbial glucoamylases: Characteristics and applications. Critical Reviews in Biotechnology, 29, 225–255.
Tanaka, A., & Hoshino, E. (1999). Study on the substrate specificity of α-amylases that contribute to soil removal in detergents. Journal of Surfactants Detergents, 2, 193–199.
Feller, G., & Gerday, C. (2003). Psychrophilic enzymes: Hot topics in cold adaptation. Nature Reviews Microbiology, 1, 200–208.
Gupta, N., Beliya, E., Paul, J. S., Tiwari, S., Kunjam, S., & Jadhav, S. K. (2021). Molecular strategies to enhance stability and catalysis of extremophile-derived α-amylase using computational biology. Extremophiles, 25, 221–233.
Cappelli, A., Oliva, N., & Cini, E. (2020). A systematic review of gluten-free dough and bread: Dough rheology, bread characteristics, and improvement strategies. Applied Sciences, 10, 6559.
Zafar, A., Aftab, M. N., Iqbal, I., ud, Din, Z., & Saleem, M. A. (2019). Pilot-scale production of a highly thermostable α-amylase enzyme from Thermotoga petrophila cloned into E. coli and its application as a desizer in textile industry. RSC Advances, 9, 984–992.
Escaramboni, B., Núñez, E. G. F., Carvalho, A. F. A., & de Oliva Neto, P. (2018). Ethanol biosynthesis by fast hydrolysis of cassava bagasse using fungal amylases produced in optimized conditions. Industrial Crops and Products, 112, 368–377.
Shad, M., Hussain, N., Usman, M., Akhtar, M. W., & Sajjad, M. (2023). Exploration of computational approaches to predict the structural features and recent trends in α-amylase production for industrial applications. Biotechnology and Bioengineering, 120, 2092–2116.
Farooq, M. A., Ali, S., Hassan, A., Tahir, H. M., Mumtaz, S., & Mumtaz, S. (2021). Biosynthesis and industrial applications of α-amylase: A review. Archives of Microbiology, 203, 1281–1292.
Gupta, R., Gigras, P., Mohapatra, H., Goswami, V. K., & Chauhan, B. (2003). J. P. b. Microbial α-amylases: A biotechnological perspective. Process Biochemistry, 38, 1599–1616.
Zinck, S. S., Christensen, S. J., Sørensen, O. B., Svensson, B., & Meyer, A. S. (2023). Importance of inactivation methodology in enzymatic processing of raw potato starch: NaOCl as efficient α-amylase inactivation agent. Molecules, 28, 2947.
Jespersen, H. M., Ann MacGregor, E., Henrissat, B., Sierks, M. R., & Svensson, B. (1993). Starch-and glycogen-debranching and branching enzymes: Prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. Journal of Protein Chemistry, 12, 791–805.
Drula, E., Garron, M. L., Dogan, S., Lombard, V., Henrissat, B., & Terrapon, N. (2022). The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Research, 50, 571–577.
Van Der Maarel, M. J., Van der Veen, B., Uitdehaag, J. C., Leemhuis, H., & Dijkhuizen, L. (2002). Properties and applications of starch-converting enzymes of the α-amylase family. Journal of Biotechnology, 94, 137–155.
Janeček, Š, Svensson, B., & MacGregor, E. A. (2014). α-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cellular and Molecular Life Sciences, 71, 1149–1170.
Stam, M. R., Danchin, E. G., Rancurel, C., Coutinho, P. M., & Henrissat, B. (2006). Dividing the large glycoside hydrolase family 13 into subfamilies: Towards improved functional annotations of α-amylase-related proteins. Protein Engineering Design and Selection, 19, 555–562.
Mareček, F., & Janeček, Š. (2022). A novel subfamily GH13_46 of the α-amylase family GH13 represented by the cyclomaltodextrinase from Flavobacterium sp. 92. Molecules, 27, 8735.
Brown, H. A., DeVeaux, A. L., Juliano, B. R., Photenhauer, A. L., Boulinguiez, M., Bornschein, R. E., Wawrzak, Z., Ruotolo, B. T., Terrapon, N., & Koropatkin, N. M. (2023). BoGH13ASus from Bacteroides ovatus represents a novel α-amylase used for Bacteroides starch breakdown in the human gut. Cellular and Molecular Life Sciences, 80, 232.
Zhou, J., Li, Z., Zhang, H., Wu, J., Ye, X., Dong, W., Jiang, M., Huang, Y., & Cui, Z. (2018). Novel maltogenic amylase CoMA from Corallococcus sp. strain EGB catalyzes the conversion of maltooligosaccharides and soluble starch to maltose. Applied Environmental Microbiology, 84, 00152–00118.
Min, B. C., Yoon, S. H., Kim, J. W., Lee, Y. W., Kim, Y. B., & Park, K. H. (1998). Cloning of novel maltooligosaccharide-producing amylases as antistaling agents for bread. Journal of Agricultural Food Chemistry, 46, 779–782.
Kim, Y. B., Yang, S. J., Lee, J. W., Cha, J., Hong, S. Y., Chung, S. H., Lee, S. T., & Rhim, S. L. (1996). Cloning and expression of an amylase gene from Streptomyces albus KSM-35 in Escherichia coli. Food Science Biotechnology, 5, 249–253.
Murakami, S., Nagasaki, K., Nishimoto, H., Shigematu, R., Umesaki, J., Takenaka, S., Kaulpiboon, J., Prousoontorn, M., Limpaseni, T., & Pongsawasdi, P. (2008). Purification and characterization of five alkaline, thermotolerant, and maltotetraose-producing α-amylases from Bacillus halodurans MS-2-5, and production of recombinant enzymes in Escherichia coli. Enzyme Microbial Technology, 43, 321–328.
Auh, J. H., Lee, S. Y., Yoo, S. S., Son, H. J., Lee, J. W., Lee, S. J., Kim, Y. B., & Park, K. H. (2005). A novel maltopentaose-producing amylase as a bread antistaling agent. Food Science and Biotechnology, 14, 681–684.
Li, Z., Wu, J., Zhang, B., Wang, F., Ye, X., Huang, Y., Huang, Q., & Cui, Z. (2015). AmyM, a novel maltohexaose-forming α-amylase from Corallococcus sp. strain EGB. Applied Environmental Microbiology, 81, 1977–1987.
Sun, Z., Li, D., Liu, P., Wang, W., Ji, K., Huang, Y., & Cui, Z. (2016). A novel L-asparaginase from Aquabacterium sp. A7-Y with self-cleavage activation. Antonie van Leeuwenhoek, 109, 121–130.
Yang, J., & Zhang, Y. (2015). I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Research, 43, W174–W181.
Qin, Y., Huang, Z., & Liu, Z. (2014). A novel cold-active and salt-tolerant α-amylase from marine bacterium Zunongwangia profunda: Molecular cloning, heterologous expression and biochemical characterization. Extremophiles, 18, 271–281.
Sanchez, A. C., Ravanal, M. C., Andrews, B. A., & Asenjo, J. A. (2019). Heterologous expression and biochemical characterization of a novel cold-active α-amylase from the antarctic bacteria Pseudoalteromonas sp. 2–3. Protein Expression and Purification, 155, 78–85.
D’Elia, J. N. (1996). Contribution of a neopullulanase, a pullulanase, and an alpha-glucosidase to growth of Bacteroides thetaiotaomicron on starch. Journal of Bacteriology, 178, 7173–7179.
Fritzsche, H. B., Schwede, T., & Schulz, G. E. (2003). Covalent and three-dimensional structure of the cyclodextrinase from Flavobacterium sp. 92. European Journal of Biochemistry, 270, 2332–2341.
Dos Santos, F. C., & Barbosa-Tessmann, I. P. (2019). Recombinant expression, purification, and characterization of a cyclodextrinase from Massilia timonae Protein Expression and Purification, 154, 74–84.
Janeček, Š. (2002). How many conserved sequence regions are there in the α-amylase family. Biologia, 57, 29–41.
Gupta, R., Gigras, P., Mohapatra, H., Goswami, V. K., & Chauhan, B. (2003). Microbial α-amylases: A biotechnological perspective. Process Biochemistry, 38, 1599–1616.
Sivaramakrishnan, S., Gangadharan, D., Nampoothiri, K. M., Soccol, C. R., & Pandey, A. (2006). α-Amylases from microbial sources–An overview on recent developments. Food Science and Biotechnology, 44, 173–184.
Song, J., Shen, X., Liu, F., Zhao, X., Wang, Y., Wang, S., Wang, P., Wang, J., Su, F., & Li, S. (2023). Ca2+-based metal-organic framework as enzyme preparation to promote the catalytic activity of amylase. Materials Today Chemistry, 30, 101522.
Fan, Q., Zhang, L., Dong, C., Zhong, L., Fang, X., Huan, M., Ye, X., Huang, Y., Li, Z., & Cui, Z. (2021). Novel malto-oligosaccharide‐producing amylase AmyAc from Archangium sp. strain AC19 and its catalytic properties. Starch‐Stärke, 73, 2100114.
Wang, J., Zhang, L., Wang, P., Lei, J., Zhong, L., Zhan, L., Ye, X., Huang, Y., Luo, X., & Cui, Z. (2023). Identification and characterization of novel malto-oligosaccharide-forming amylase AmyCf from Cystobacter sp. Strain CF23. Foods, 12, 3487.
Wu, J., **a, B., Li, Z., Ye, X., Chen, Q., Dong, W., Zhou, J., Huang, Y., & Cui, Z. (2015). Molecular cloning and characterization of a novel GH13 saccharifying α-amylase AmyC from Corallococcus sp. EGB. Starch‐Stärke, 67, 810–819.
Li, S., Zuo, Z., Niu, D., Singh, S., Permaul, K., Prior, B. A., Shi, G., & Wang, Z. (2011). Gene cloning, heterologous expression, and characterization of a high maltose-producing α-amylase of Rhizopus oryzae. Applied Biochemistry Biotechnology, 164, 581–592.
Dauter, Z., Dauter, M., Brzozowski, A. M., Christensen, S., Borchert, T. V., Beier, L., Wilson, K. S., & Davies, G. J. (1999). X-ray structure of Novamyl, the five-domain maltogenic α-amylase from Bacillus stearothermophilus: Maltose and acarbose complexes at 1.7 Å resolution. Biochemistry, 38, 8385–8392.
Kim, J. S., Cha, S. S., Kim, H. J., Kim, T. J., Ha, N. C., Oh, S. T., Cho, H. S., Cho, M. J., Kim, M. J., & Lee, H. S. (1999). Crystal structure of a maltogenic amylase provides insights into a catalytic versatility. Journal of Biological Chemistry, 274, 26279–26286.
Qi, Y., Geib, T., & Volmer, D. A. (2015). Determining the binding sites of β-cyclodextrin and peptides by electron-capture dissociation high resolution tandem mass spectrometry. Journal of the American Society for Mass Spectrometry, 26, 1143–1149.
An, Y., Tran, P. L., Yoo, M. J., Song, H. N., Park, K. H., Kim, T. J., Park, J. T., & Woo, E. J. (2023). The distinctive permutated domain structure of periplasmic α-amylase (MalS) from glycoside hydrolase family 13 subfamily 19. Molecules, 28, 3972.
Chen, X., Zhang, L., Li, X., Qiao, Y., Zhang, Y., Zhao, Y., Chen, J., Ye, X., Huang, Y., & Li, Z. (2020). Impact of maltogenic α-amylase on the structure of potato starch and its retrogradation properties. International Journal of Biological Macromolecules, 145, 325–331.
Zhang, L., Zhong, L., Wang, P., Zhan, L., Yangzong, Y., He, T., Liu, Y., Mao, D., Ye, X., & Cui, Z. (2023). Structural and functional properties of porous corn starch obtained by treating raw starch with AmyM. Foods, 12, 3157.
Zhang, L., Li, Z., Qiao, Y., Zhang, Y., Zheng, W., Zhao, Y., Huang, Y., & Cui, Z. (2019). Improvement of the quality and shelf life of wheat bread by a maltohexaose producing α-amylase. Journal of Cereal Science, 87, 165–171.
Yang, T., Zhong, L., Jiang, G., Liu, L., Wang, P., Zhong, Y., Yue, Q., Ouyang, L., Zhang, A., & Li, Z. (2022). Comparative study on bread quality and starch digestibility of normal and waxy wheat (Triticum aestivum L.) modified by maltohexaose producing α-amylases. Food Research International, 162, 112034.
Kelly, C. T., Collins, B. S., Fogarty, W. N., & Doyle, E. M. (1993). Mechanisms of action of the α-amylase of Micromonospora melanosporea. Applied Microbiology Biotechnology, 39, 599–603.
Collins, B. S., Kelly, C. T., Fogarty, W. M., & Doyle. (1993). The high maltose-producing α-amylase of the thermophilic actinomycete, Thermomonospora curvata. Applied Microbiology and Biotechnology, 39, 31–35.
Doyle, E. M., Kelly, C. T., & Fogarty, W. M. (1989). The high maltose-producing α-amylase of Penicillium Expansum Applied Microbiology Biotechnology, 30, 492–496.
McMahon, H. E., Kelly, C. T., & Fogarty, W. M. (1999). High maltose-producing amylolytic system of a Streptomyces Sp. Biotechnology Letters, 21, 23–26.
Funding
This work was supported by the Natural Science Foundation of China (32370119) and Liaocheng University (No 318052291).
Author information
Authors and Affiliations
Contributions
YX Wang and XF Ye designed the experiments and wrote the manuscript. YX Wang carried out the experiments. HR Xue and ZS Zhao provided help in the construction of the plasmid. TT **e and GH Yan helped to finish the experiments. XF Ye provided valuable suggestions for improving the experimental methods. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., **e, T., Yan, G. et al. Heterologous Expression and Characterization of a Novel Mesophilic Maltogenic α-Amylase AmyFlA from Flavobacterium sp. NAU1659. Appl Biochem Biotechnol (2024). https://doi.org/10.1007/s12010-024-04874-x
Accepted:
Published:
DOI: https://doi.org/10.1007/s12010-024-04874-x