Abstract
Electroencephalogram (EEG) is one of most effective clinical diagnosis modalities for the localization of epileptic focus. Most current AI solutions use this modality to analyze the EEG signals in an automated manner to identify the epileptic seizure focus. To develop AI system for identifying the epileptic focus, there are many recently-published AI solutions based on biomarkers or statistic features that utilize interictal EEGs. In this review, we survey these solutions and find that they can be divided into three main categories: (i) those that use of biomarkers in EEG signals, including high-frequency oscillation, phase-amplitude coupling, and interictal epileptiform discharges, (ii) others that utilize feature-extraction methods, and (iii) solutions based upon neural networks (an end-to-end approach). We provide a detailed description of seizure focus with clinical diagnosis methods, a summary of the public datasets that seek to reduce the research gap in epilepsy, recent novel performance evaluation criteria used to evaluate the AI systems, and guidelines on when and how to use them. This review also suggests a number of future research challenges that must be overcome in order to design more efficient computer-aided solutions to epilepsy focus detection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epilepsy, one of the most common neurological disorders, can affect people of any age, race, or ethnic background. According to the latest study by the World Health Organization (WHO), approximately 65 million people worldwide are affected by epilepsy, and there are an estimated 2.4 million new cases each year (Giannakakis et al. 2014; Levesque et al. 2017; Stafstrom and Carmant 2015). Epilepsy, defined as repeated and unpredictable seizures, causes social impairment and a high risk of death (Fisher et al. 2014; Pati and Alexopoulos 2010). Childhood epilepsy also seriously impacts the development of the brain by reducing learning ability and mental growth. Epileptologists generally classify seizures as either focal or generalized based on abnormal brain activities (Ngugi et al. 2011; van Mierlo et al. 2014). To control epileptic seizures, epileptologists prescribe anti-epileptic drugs. When these medicines fail to control the seizures, surgical removal of the epileptic focus may be the patient’s best chance for seizure freedom.
According to the clinical guidelines related to epilepsy surgery, the epileptic seizure focus is the cortex area from which the seizures originate (Lüders et al. 2006). The identification and surgical removal of the focus must be resected (inactivated or completely disconnected) for complete seizure freedom. Standard diagnostic methods include inspection of seizure semiology, high-resolution magnetic resonance imaging (MRI), and EEG. The scalp EEG is a non-invasive method of recording electrical activity by placing electrodes on the scalp using the international standard 10–20 system (Paul 2018). It provides one of the promising ways to identify the epileptic seizure focus before surgical intervention. However, when epileptologists cannot determine an epileptic seizure focus using non-invasive methods, they indicate to use intracranial EEGs (iEEGs) with the implantation of intracranial electrodes during both interictal and ictal phases. Before the epileptic focus resection for complete abolition of seizures, the epilepsy surgeon should consider integrating the multi-channel intracranial electrodes to these “area of cortex” and recording iEEG signals until collecting enough data from habitual seizures to analyze. The epileptologists then need to inspect the multi-channel iEEG data to identify seizure onset zones (SOZs) from within the recorded cortex area. During these inspections, the epileptologists need to analyze and label all long-term multi-channel iEEG data, the manual detection of which is challenging and time-consuming. The success of the epileptic focus resection for seizure freedom depends on accurate detection of the seizure focus. A key to achieving good results with resection surgery is identifying and resecting the area that may cause seizures. Such areas are called epileptogenic zones (EGZs). Currently, there is no single, non-invasive test method that can identify those areas. Approximately 20–30 percent of patients suffer from recurrent seizures after surgery (Elsharkawy et al. 2011).
In the endeavor to design computer-aided diagnosis tools, both non-invasive and invasive iEEGs are promising procedures. Automatic detection of seizure focus is highly desired, as it would reduce the epileptologist’s workload and would, along with other tests, increase confidence in related medical decisions. Also, computer-aided and data-driven approaches may provide a way of revealing a mechanism of epileptogenesis.
For designing the computer-aided systems based on the different types of EEG modalities, Some recent studies used biomarkers, including high-frequency oscillations (HFOs)(Zijlmans et al. 2011; Jacobs et al. 2009; Urrestarazu et al. 2007), phase-amplitude coupling (PAC)(Guirgis et al. 2015; Motoi et al. 2018; Amiri et al. 2019), interictal epileptiform discharges (IEDs) (Staley and Dudek 2006; Elsharkawy et al. 2011) while others utilized feature-extraction methods (Sharma et al. 2015b; Akter et al. 2020a, 2019; Itakura and Tanaka 2017). In biomarker-related studies to identify epileptic seizure focus, the computer-aided solutions have combined the epilepsy biomarkers in EEG signals with advanced signal and machine-learning approaches. The epilepsy biomarkers in EEGs are essential for identifying the epileptic seizure focus within conventional clinical systems. However, from a machine learning perspective, the features extracted from EEG signals to characterize epileptic signals are also good candidates for identification of epileptic focus. To identify the epileptic focus from EEG signals, the recorded EEG signals are typically pre-processed using various signal processing methods, and features are extracted from the pre-process EEGs signals to represent in a compact form (Akter et al. 2020a, 2019; Hassan et al. 2019). The different studies introduced feature-extraction methods to design the computer-aided solutions for identifying epileptic focus (Sharma et al. 2015b; Akter et al. 2020a, 2019; Hassan et al. 2019; Itakura and Tanaka 2017). Based on the biomarker and feature-extraction related studies, there is a large diversity of research that have been explored in the last years to design AI-systems. Many new algorithms have been designed and explored in order to identify epileptic seizure focus.
However, there are few review papers that surveys works on automated machine learning-based focus detection methods. Recently, one review paper for focal and non-focal epilepsy localization (Hussein et al. 2018) has summarized the several studies with a variant of datasets to identify focal and non-focal channels. However, the main goal of most studies reported in their survey was to identify seizure patterns in continuous EEG signals instead of localizing focal and non-focal channels used in Bern Barcelona dataset (Andrzejak et al. 2001). For diverse research in the epilepsy, we would like to draw attention to the AI-solutions that different researchers have introduced for identifying the epileptic focus. In this paper, we survey the literature of the past decade to observe which new approaches have been investigated to design computer-aided automated systems and be the most efficient.
Epileptic seizure focus
The epileptogenic zone (EGZ) is a conceptual definition of the area that causes seizures; resection of the EGZ halts them. When antiepileptic drugs fail to treat epilepsy, the patient is diagnosed with refractory epilepsy. One of the treatment approaches is removing the cortex’s possible area responsible for generating the abnormal neural activity resulting in epileptic seizure. In a study of surgical treatment of epilepsies, Lüders et al. (2006) categorized the EGZ as actual EGZ (commonly referred to SOZ) and potential EGZ. The study defined the SOZ as the cortex area from which actual clinical seizures are generated. The SOZ is most commonly localized by either scalp or intracranial EEG. In contrast, after resecting the SOZ, the seizure may continue from the other area of the cortex defined as the potential SOZ. This situation occurs when the surgery does not accomplish complete resection of the actual EGZ.
So far, the precise localization of the EGZ boundary cannot be characterized by any diagnostic methods. The most common methods to localize the SOZ are either scalp EEG or iEEG during both the interictal and ictal phases. Varied recent epilepsy research has suggested the use of ictal and interictal high-frequency activity (\(\ge 80\) Hz) to localize the epileptogenic zone. In clinical practice, the SOZ is used to localize the boundary of the EGZ (Rosenow and Lüders 2001). In defining the possible boundary of the EGZ, the different cortical zones are measured using some standard diagnosis methods. The descriptions of these cortical zones and diagnostic tests are provided in Table 1. During the presurgical evaluation, epileptologists should consider these cortical zones when estimating the location and extent of EGZ (Talairach and Bancaud 1966; Lüders et al. 2006). To localize the possible EGZ, Talairach and Bancaud (1966) introduced three zones, namely a lesional, an irritative, and an epileptogenic zone. They reported that the SOZ was a reliable index of the location and extent of the possible EGZ and defined the SOZ as the epileptogenic zone.
Decades later, Lüders et al. (2006) enriched that concept by defining five cortical zones: the irritative zone (generates interictal spikes), seizure-onset zone (initiates clinical seizures), the symptomatogenic zone, the lesion, and the functional deficit zone. The detailed presurgical evaluation always requires defining these zones to understand the spatial relationship with the epileptogenic zone. Lüders et al. (2006) observed that, for some patients, complete resection of the SOZ does not lead to seizure freedom. After resecting the SOZ, the epileptic seizures were generated from the area in the brain closest to SOZ. Their study further dichotomized the SOZ into the actual and potential SOZ and suggested that the complete resection of both the actual and potential SOZ may be necessary for complete seizure cessation (see in Fig. 1). Therefore, epilepsy surgeons and epileptologists need to define the location and the extension of the EGZ by observing both SOZs before epileptic focus resection. Since the location and extension of the EGZ is a theoretical concept based on defining the actual and potential SOZs, clinical experts may only decide about the EGZ if the patient after surgery is seizure-free. Bancaud and Talairach (1992) also observed that, with patients with medically-intractable epilepsy, the SOZ is involved in a single region and interconnected with distinct regions.
Research is still ongoing regarding identification of the SOZ and the extent of brain area that should be considered for resection in order to achieve successful surgical treatment of epilepsy. In conventional clinical procedure for identification of the SOZ ( both actual and potential), the EEGs with multiple channels are recorded until enough data on habitual seizures has been collected. An epileptologist visually inspects the long-term multi-channel EEG signals to identify the electrodes with the possible SOZ that must be resected in order to achieve freedom from seizures. However, this process is challenging and time-consuming for epileptologists. Several studies have recently proposed computer-aided solutions based on either epilepsy biomarkers (HFOs, PAC, and IEDs) or the use of feature-extraction methods in EEGs. In the rest of this paper, we have reviewed the computer-aided designs (CAD) that might be utilized to identify the possible SOZ (Table 2).
Public datasets
Public epilepsy datasets can be roughly divided into two categories: scalp EEG and intracranial EEG. The first is used to detect abnormalities related to epileptic seizure and the second to determine epileptic focus location. What follow are some of the most commonly-used scalp EEG datasets.
-
The Temple University Hospital (TUH) EEG Corpus (Harati et al. 2014), which is a large size and contains various sub-datasets, including abnormality detection, seizure detection, and artifact classification.
-
The Children’s Hospital Boston (CHB-MIT) Scalp EEG Dataset (Goldberger et al. 2000) (Shoeb 2009), which is used for seizure detection.
-
The University of Bonn EEG Dataset (Andrzejak et al. 2001) contains several different classes of data that are recorded from healthy volunteers and patients.
-
Other common datasets include IEEG.org and the European Epilepsy Dataset.
Regarding the epileptic focus location problem, the most used public intracranial EEG dataset is the so-called Bern-Barcelona EEG dataset (Andrzejak et al. 2012). This dataset includes the iEEG signal recorded from five patients with long-standing pharmacoresistant temporal lobe epilepsy who are candidates for epilepsy surgery. iEEG signals are recorded by the AD-TECH device (Racine, WI, USA), which has a sampling rate of 512 Hz. The focal channel, which detected first ictal iEEG signal changes and judged throw visual by at least two clinical experts. The other channels are defined as non-focal channels. An example of focal and non-focal samples are shown in Fig. 2. The dataset includes 7500 samples of the focal and non-focal, respectively. Each sample is 20 seconds and is processed with the bandpass filter between 0.5 and 150 Hz by using fourth-order Butterworth filter.
This dataset has been extensively used as a benchmark in the literature; however, it also has the following drawbacks:
-
It includes no information regarding locations of electrodes, which is essential for focal identification.
-
Signals are provided as independent segments without patient labels.
-
The highest frequency is limited to 150 Hz, even though recent neurological findings indicate that high frequency components (>100 Hz) are crucial to identify the epileptic focus.
Bio-marker-based approach
High frequency oscillation
High-frequency oscillations (HFOs >80 Hz) are short durational high gamma ripple and fast ripple activities in EEG signals, which are one of the biomarkers used to identify epileptic focus in conventional clinical settings (Zijlmans et al. 2011; Jacobs et al. 2009; Urrestarazu et al. 2007). In the human intracranial EEG signal, the frequency components of HFOs are greater than 80 Hz and can range up to 500 Hz. The sampling frequency needs to be at least 10 kHz to record HFOs in EEG signals. In general, the amplitude of HFOs varies between 10 and 1000 \(\mu \)V. An example of a filtered electrocorticography (ECoG) signal in the ripple band (80–250 Hz) and in a low frequency range (0.5–60 Hz) are shown in Fig. 3. To analyze the HFOs, extra care is necessary, as they might be corrupted by broad frequency components, such as spiking activity or other artifacts (Worrell et al. 2012). The HFOs are commonly categorized as ripples (100–250 Hz) or fast ripples (250–500 Hz) and are generally specific to the SOZ (Crépon et al. 2010). They are crucial for epilepsy surgery for removing entirely epileptic seizure focus required to produce seizure freedom (Lüders et al. 2006).
Example of filtered ECoG signal in the low frequency range of (0.5–60 Hz) and in the ripple band (80–250 Hz). A16–A23 are the channel names. a is the sub-figure of the filtered ECoG signal for 10s in a low frequency range of (0.5–60 Hz). b is the filtered ECoG signal for 1s in the ripple band (80–250 Hz)
Automated methods for HFOs detection
The methods of identification of HFOs are classified as manual, automatic, and a combination of the two approaches (Navarrete et al. 2016a). Manual detection, carried out by an expert or panel of experts, consists of visually monitoring a computer display. Experts must have a thorough understanding of electrophysiological recording and signal processing. Due to the current exponential growth in multi-channel measurement, manual inspection has become a difficult process, a situation that has undoubtedly increased the need for automatic detection. Therefore, to detect HFOs, several studies have proposed automated detectors over the past decade.The methodological advances for detecting HFOs in EEG signals occurred between 2002 and 2016 and Navarrete et al. (2016b) summarized in their study.
In the design of the HFO-based automated system, the main objective was to detect putative HFOs, including the pathological HFOs (pHFOs) and physiological-induced HFOs (nHFOs). The nHFOs, which were rejected based on manual observation by experts (Staba et al. 2002; Zelmann et al. 2012; Bénar et al. 2010; Liu et al. 2016) or implementation of complex methods based on machine-learning techniques (supervised or unsupervised) (Blanco et al. 2010; Matsumoto et al. 2013; Chaibi et al. 2014; Sciaraffa et al. 2020). There are many ways to extract putative HFOs from the desired frequency bands. The most common types of methods for automated detection are energy-thresholding-based methods (Staba et al. 2002; Zelmann et al. 2012; Bénar et al. 2010), including short-time energy (STE) (Staba et al. 2002), time line-length energy (Gardner et al. 2007), Hilbert transform envelope (HTE) (Crépon et al. 2010), the Montreal Neurological Institute (MNI) detector (Zelmann et al. 2012). Birot et al. (2013) combined three approaches, root mean square (RMS) amplitudes, line-length energy (LLE), and instantaneous frequency (IF), to identify HFOs. The above four methods for automated HFOs detection were also implemented in RIPPLELAB to assess performance (Navarrete et al. 2016a). Subsequently, other groups have proposed and tested algorithms, including Bumps modeling techniques (BMT) (Chaibi et al. 2013), estimation of power spectra by multitaper methods (Wang et al. 2014), RSM amplitudes in referential montage (Gliske et al. 2016), baseline detection by wavelet entropy (WE), and Hilbert envelope (Fedele et al. 2016), to monitor HFOs.
In order to deal with the classification problem of possible HFO events, the supervised and unsupervised classifiers such as clustering, Gaussian mixture model (GMM), support vector machines (SVM), random forest classifier (RF) are the useful method to differentiate the physiological and pathological HFOs (Blanco et al. 2010; Matsumoto et al. 2013; Chaibi et al. 2014; Sciaraffa et al. 2020; Varatharajah et al. 2018). However, classifiers’ unsupervised adaptation is much more difficult than supervised adaptation, as the class labels are unknown. The first examples of unsupervised approach (K-mediods clustering) were applied to detect HFO events proposed by Blanco et al. (2010). In modern clinical practice, the clinical translation of HFOs as a biomarker of EEGs has been largely restricted due to the difficulty in differentiating pathological HFOs from physiological HFOs (Jacobs et al. 2018; Fedele et al. 2019). Supervised automated detection techniques with SVM classifier were proposed by Matsumoto et al. (2013) for the differentiation between physiological and pathological HFOs. The line-length energy (LLE) method was used to a sliding 200-ms window with 50-ms overlap to detect the pathological HFOs (pHFOs) and physiological-induced HFOs (nHFOs) from background EEGs. A SVM classification was performed on the pHFO and nHFO using a rotating cross-validation approach by training the data for four patients and then classifying the remaining patient data. Chaibi et al. (2014) used six feature extraction methods to design an automated system. In the design of an automated system, a decision tree (DT) classifier has been trained with features of 200 events (100 events of background activity and 100 events of HFOs activity). In the testing phase, the 50-ms segments with with overlap** one sample were used to mark by 1 over time. They compared their system with five energy-based different approaches (STE (Staba et al. 2002), complex morlet wavelet (CMW) (Khalilov et al. 2005), Bumps modelling (BM) technique (Chaibi et al. 2013), and Hilbert (Zelmann et al. 2012)). Subsequently, different classification methods including linear discriminant analysis (LDA) (Jrad et al. 2015), SVM (Jrad et al. 2017), and radial basis function SVM (RBF-SVM) have been tested to detect HFOs in several studies (Sciaraffa et al. 2020). Some recent studies also try to adapt deep learning-based approaches for identifying the two kinds of HFOs in ripple and fast ripple iEEG (Medvedev et al. 2019; Zuo et al. 2019; Firpi et al. 2007). However, the above studies mainly focus on detecting HFOs in ripple and fast ripple iEEG data in a separate way rather than identifying the epileptic focus.
Phase-amplitude coupling (PAC)
Many studies have recently reported on phase-amplitude coupling (PAC) analysis of epileptic EEG. Nariai et al. (2011) have observed that the interictal HFOs at 80–200 Hz within the seizure onset zone (SOZ) are tightly locked to the phase more than 3 Hz of slow-wave. On the contrary, ictal HFOs are tightly locked to the phase less than 1 Hz of slow-wave. Guirgis et al. (2015) investigated that low-frequency oscillation (LFO)-modulated HFOs might be employed to identify regions of interest (ROI) in extratemporal lobe patients. Moreover, Amiri et al. (2016) found that PAC was more substantial in the SOZ than outside of it, and PAC was higher in spiking channels outside the SOZ than in normal brain areas. Another study found that the HFOs coupled to phases of theta band can be used to identify SOZ (Amiri et al. 2019). Weiss et al. (2016) examined the ripple amplitude, which was significantly modulated by a phase of epileptic EEG spike with the SOZ. Last, Motoi et al. (2018) reported in their study that PAC strength is different between the ripple and a phase of 3–4 Hz.
PAC can be simply explained as the coupling on high-frequency amplitude modulated by low-frequency phase, which reveals the coupling phenomenon between brain rhythms, especially between slow-waves and fast-waves, like that seen in high-frequency oscillation (HFO). In many studies, the PAC strength is quantified using the mean vector length modulation index (MVL-MI) (Canolty et al. 2006). Specifically, there are three steps to calculate the PAC strength through the MVL-MI approach. Firstly, defining a complex component as
where the high-frequency amplitude and low-frequency phase are denoted as \(A_{amp}(t)\) and \(\phi _{pha}(t)\), respectively. Secondly, defining the raw modulation index as
Thirdly, a surrogate data approach is introduced to z-scored the \(MI_{raw}\) by using the mean (denoted as \(\mu \)) and standard deviation (denoted as \(\sigma \)) of 100 surrogate data. The final modulation index can then be defined as
where the MI is used to evaluate PAC strength.
Hereafter, in order to graphically display a series of PAC values of different frequencies, the phase-amplitude comodulogram approach is introduced. A simple example is shown in Fig. 4, the frequency of phase is represented in the horizontal axis, while the frequency of amplitude is displayed in the vertical axis. The PAC value between the designated frequency of phase and the amplitude’s designated frequency is exhibited using a pseudocolor plot in which a hot color denotes high coupling strength.
In more recent studies, some machine-learning methods have been applied to identification of the SOZ based on PAC features. Varatharajah et al. (2018) proposed the support vector machine (SVM) approach with PAC features to detect SOZ. Elahian et al. (2017) employed logistic regression technology based on the PAC values of ictal ECoG recordings to identify SOZ electrodes. In the last decade, it has become popular to combine promising features and AI-based models for SOZ detection. Table 3 summarizes the literature about PAC analysis for SOZ localization that was published between 2015 and 2019. ETLE, MTLE, and mTLE represent extratemporal lobe epilepsy, mesial temporal lobe epilepsy, and mesiotemporal lobe epilepsy, respectively. \( MI_{30-450 Hz \& 0.5-4 Hz}\) denotes PAC strength between the amplitude of 30–450 Hz and the phase of 0.5–4 Hz.
Interictal epileptiform discharges (IEDs)
Interictal epileptiform discharges (IEDs), which are generated from the irritative zone, are another important biomarker of epilepsy surgery that is used to localize or extend epileptic focus. The IEDs are morphologically classified as spikes, sharp waves, poly-spike complexes, or as a multiple-spike complex (Noachtar et al. 1999; Staley and Dudek 2006; De Curtis and Avanzini 2001). The IEDs are clearly distinguished from background activity through visual inspection of long-term multichannel EEGs by EEG experts. Figure 5, which was collected from the Juntendo Hospital, shows an example of IEDs (blue circle) in the SOZ channels. According to the definition set by the International Federation of Societies for Electroencephalography and Clinical Neurophysiology (IFSECN) (Deuschl 1999; De Moraes and Callegari 2014), the spikes have a duration between 20 and 70 ms, while sharp waves last between 70 and 200 ms. The spike-and-wave complexes are defined as spikes followed by slow waves that have a duration from 150 to 350 ms. However, standard modalities to observe the IEDs from background activity are either EEG or MEG recorded from epilepsy patients (Lüders et al. 2006). Since our review study is primarily concentrated on detecting epileptic focus using EEG signals, most of the algorithms that have been introduced in this section based on EEG spike detection methods. It is important to note that detection of IEDs in the EEG signals is one of the most difficult tasks for epileptologists due to the variation of spikes morphology and their similarities to artifacts (Wilson and Emerson 2002; Oikonomou et al. 2007) such as muscle activity and eye blinks.
In automated IED-based detection studies, many detection algorithms have been proposed based on single and multichannel EEG approaches (Oikonomou et al. 2007). The reviews that study automatic detection of IED spikes have been summarized in several review papers over the last four decades. (Pardey et al. 1996; Wilson and Emerson 2002; Abd El-Samie et al. 2018; Halford 2009; De Moraes and Callegari 2014). Several studies have categorised the existing detection methods into mimetic analysis (Gotman and Gloor 1976; Exarchos et al. 2006; Boos et al. 2011; Ji et al. 2011b; Halford et al. 2013), template matching (Stevens et al. 1972; Fischer et al. 1980; Kalayci and Ozdamar 1995; Ji et al. 2011a; Nonclercq et al. 2012), parametric approaches (Oikonomou et al. 2007; Durka 2004; Acır and Güzeliş 2004; Radmehr and Anisheh 2013),and power spectral analysis (Adjouadi et al. 2004a; Barlow 1980). The mimetic approaches to detection of IEDs from background activity split the long-term iEEG signals into half-waves based on amplitude extrema; two half-waves create the triangular shape of an epileptic transients with opposite directions (Halford 2009). In identifying the waves related to IEDs, each wave is examined against the set of predetermined criteria provided by EEG experts. These criteria were created based upon the distinctive attributes of IEDs, namely area, slope, amplitude, duration, sharpness, and dominant frequency. Most of the detection methods are based on thresholding (Garg and Kohli 2013; Juozapavicius et al. 2011; Xu et al. 2007, 2006) or on complex algorithms with a classifier (Exarchos et al. 2006; Saastamoinen et al. 1998; Liu et al. 2013). Nevertheless, as many studies have noted, the traditional morphological definition can be changed, particularly during long-term iEEG recording due to eye blinks, vertex waves, electrode artifacts, and movement artifacts (Adjouadi et al. 2004a; Nakamura et al. 2001). To improve the performance of the mimetic approach, some studies have utilized pre-processing via the the Walsh operator, Fourier transform (FT), or incorporation of additional channels, such as electrocardiogram (ECG), electrooculogram (EOG), or electromyogram (EMG) signals (Glover et al. 1989; Black et al. 2000; Nakamura et al. 2001).
Biomedical signals like EEGs are mostly non-stationary, which implies a time-varying frequency spectrum. For such signals, either an adaptive or a non-adaptive model can be used to identify epileptiform events (Isaksson et al. 1981; Pardey et al. 1996; Oikonomou et al. 2007). Some attempts have been made to describe EEG signals through use of a mathematical model characterized by a set of parameters. Methods, such as autoregressive (AR), moving average (MA), and autoregresssive moving average (ARMA), can be utilized to create time-invariant models for EEG signals. Under the assumption of non-stationarity epileptiform events, Kalman’s filtering approaches have also been adapted to enhance EEG signals before spike detection (Oikonomou et al. 2007). However, these methods are prone to false detections due to contamination or corruption by a set of artifacts (Kalayci and Ozdamar 1995; Azami and Sanei 2014; Oikonomou et al. 2007). For recent advances in AI-based methodologies, some authors have combined advanced signal processing approaches and classification with the above techniques to isolate IED spikes from background signals. The wavelet transform is a powerful tool in signal processing that has led to numerous biomedical applications, and several studies have focused on its use in neural networks to detect EEG spikes (Halford et al. 2013; Song and Zhang 2013; Kalayci and Ozdamar 1995; Gopan et al. 2013; Chavakula et al. 2013). Azami and Sanei (2014) used smoothing techniques with noisy scenarios, namely, discrete wavelet transform (DWT), Kalman filter (KF), singular spectrum analysis (SSA), Savitzgy-Golay (SG) filter, and empirical mode decomposition (EMD) for improving performance of AI-solution.
Many studies have focused on develo** template-matching approaches (Lodder et al. 2013; Ji et al. 2011b, a; El-Gohary et al. 2008; Hese et al. 2008) in which the EEG experts visually select a few spikes from a set of EEG signals. These spikes were averaged to generate a template, and time-shifted correlation approaches were applied to determine the similarity between each segment and the template. Automated techniques to identify the spikes include simply checking the threshold or the use of algorithms, such as clustering, SVM, or more complex classifiers (Lodder et al. 2013; Ji et al. 2011b, a; El-Gohary et al. 2008; Hese et al. 2008). A frequency-domain analysis can be an important tool in the analysis of epileptic spikes. The most common power spectral analyses are used in epileptic spike detection, including Fourier transform (FT) (Logesparan and Rodriguez-Villegas 2011), Hilbert transform (HF) (Witte et al. 1991; Feucht et al. 1997), Walsh transform (WT) (Adjouadi et al. 2004b), and more advanced methods, such as wavelet analysis (Özdamar and Kalayci 1998; Goelz et al. 2000). In order to develop a more accurate detection system, some studies have proposed decomposition methods to divide the EEG signals into narrow subbands (Bourien et al. 2004; Pietilä et al. 1994; Witte et al. 1991), such as delta, theta, alpha, beta, and gamma, which allows for accurate estimation of the distinctive attributes of interictal spikes. Periodicity-based methods may be performed because EEG signals may exhibit some periodicity in the presence of IED spikes (Fürbass et al. 2015; Herta et al. 2015; Koren et al. 2015). In recent years, deep learning techniques like convolutional neural networks (CNN) have exhibited superior performance regarding detection of IEDs in EEG signals, especially when trained on large databases (Fukumori et al. 2019). However, because the detection of IEDs is critical to extending the boundary of SOZ, IED-related research has mainly focused on identifying IEDs rather than on localization of epileptic focus detection (Table 4).
Statistical feature extraction
Apart from the above biomarker-related, computer-aided design (HFOs, PAC, and IEDs), the statistical features have, over the last couple of years, also been used as an engineering markers for the identification of possible SOZ. It is important to note that the iEEG is the biosignals, and such biosignals contain useful information of diseases about biological systems. However, it is not easy to identify that information solely through visual observation of the raw signals. EEG signals have a significantly low signal-to-noise ratio (SNR) and are frequently corrupted during recording by external- and internal-source artifacts (Islam 2015; Islam et al. 2018). The presence of artifacts and noise poses a significant challenge to analysis of recorded signals, and thus, to useful information extraction or classification. The direct use of machine-learning approaches to biosignals without pre-processing or feature extraction may lead to a decrease in sensible decisions by the automated system. Pre-processing, signal processing, and machine-learning approaches were combined with a standard computer-aided design to deal with EEG artifacts and to extract and decode the relevant features of EEG signals. In an epileptic focus detection system, the most commonly used pre-processing step was division of the long-term EEG data into a set of segments or frames. The segmentation of that long-term EEG data was dependent upon data size, system performance, and the direction of clinical experts (Akter et al. 2020a, 2019; Hassan et al. 2019).
Feature-extraction methods can be applied to the segments to extract the practical features. To implement epileptic focus detection, several studies since 2020 have suggested utilizing information-theoretic and statistical approaches. The statistical features have been shown to be particularly useful at detecting the epileptic focus in different studies. After extracting features from iEEG signals, the next step is to classify the normal and epileptic events using a classifier, such as k-nearest neighbor (kNN), linear discrimination analysis (LDA), support vector machine (SVM), or an implementation of a complex algorithms (i.e., neural networks (NN)).
For the design of computer-aided solutions to the problem of identifying epileptic seizure focus, several epilepsy research exploited Bern-Barcelona iEEG dataset, which was considered benchmark (Table 5). The Bern-Barcelona iEEG dataset consists of a pair of focal and non-focal signals recorded in the epileptogenic and non-epileptogenic zones of the brain. In order to observe the focal and non-focal channels in iEEG, Zhu et al. (2013) first introduced the delay permutation entropy (DPE) method with an SVM classifier. Experimental results show that the DPE index of focal channels was significantly lower than that of the non-epileptogenic hemisphere. Sharma et al. (2014) proposed sample entropy as a feature and the least square SVM (LS-SVM) classifier was used to discriminate focal and non-focal features. The mean and standard deviation (SD) were also used to differentiate between the two channels (Deivasigamani et al. 2016). Three machine-learning approaches, including kNN, fuzzy Sugeno classifier (FSC), and LS-SVM, have been trained using the wavelet entropy features (Sharma et al. 2015b). The features were Shannon, Renyi’s, Tsallis, fuzzy, permutation, and phase entropies. However, the nature of EEG and ECoG signals is very complex, non-stationary, and time-dependent. Due to the non-stationary properties of EEG data, the linear feature-extraction methods do not adapt perfectly to the EEG signals. Therefore, several studies proposed decomposition methods to improve the performance of the system, including wavelet transformation (WT) (Gupta and Pachori 2020), wavelet filter-bank (WFB) (Sharma et al. 2017), tunable-Q wavelet transformation (TQWT) (Sharma et al. 2017), flexible analytic wavelet transform (FAWT) (Gupta et al. 2017), and Empirical WT (EWT) (Bhattacharyya et al. 2018). A comparison study with Bern-Barcelona dataset using time-domain multiband analysis, including EMD and bivariate EMD (BEMD) (Rilling et al. 2007; Subasi et al. 2019), was reported by Itakura and Tanaka (2017) and (Bhattacharyya et al. 2018). Recently, new developments in computer-aided systems have suggested 23 feature-extraction methods that might be used to extract features from EEG signals (Acharya et al. 2019). The feature-extraction methods used to design those systems were different types of entropies, fractal dimension (FD), RMS, Hjorth parameters, Hurst exponent, KLC, LZC, and QRA. Detailed descriptions of the feature-extraction methods are provided in Table 4. To correctly detect focal and non-focal EEG signals, LS-SVM was adapted with feature ranking based on student t-test. To demonstrate the efficacy of features, some authors have investigated the several kinds of classifiers, including adaptive neuro fuzzy inference system (ANFIS), probabilistic neural network (PNN), naïve bayes (NB), non-nested generalized exemplars classifier (NNge), best first decision tree (BFDT), and radial basis function kernel-SVM (RBF-SVM). Though most of the research performed since has used the Bern-Barcelona dataset to design computer-aided systems capable of detecting epileptic seizure focus, some have explored different feature-extractions and machine-learning methods. Algorithmic progress has been limited due to the lack of a standard multi-channel benchmark iEEG datasets. Nevertheless, how the implementation of any of these methods will influence epileptic focus detection for real clinical applications is not yet clear.
Recently neurology hospitals measure ECoG with a high sampling frequency (20 kHz) and high-density electrode grids (multi-channels) in order to localize the epileptic seizure focus. Most recently, the connectivity and synchronization processes of the epileptic brain have been analyzed in different studies (Klimes et al. 2015; Warren et al. 2010; Antony et al. 2013). In general, the epileptic brain is distinguished by increased neuronal synchrony, and the epileptic focus is often functionally disconnected from the surrounding areas (Warren et al. 2010; Antony et al. 2013). The energy variation of the SOZ is significantly higher than that of the non-epileptic zone of the ripple (80–250 Hz), and fast ripple (250–600 Hz) bands (Klimes et al. 2015; Warren et al. 2010; Antony et al. 2013). In particular, the repetitive waveform pattern called the high-frequency oscillations (HFOs) consists of ripple, and fast ripple are the valid frequency bands to localize the epileptic seizure focus (Jacobs et al. 2009; Zijlmans et al. 2011; Urrestarazu et al. 2007). Based on these recent findings related to epileptic focus and clinical application, the computer-aided solutions that utilize feature-extraction methods encompass several open research questions that deserve to be addressed. The Bern-Barcelona dataset does not support the neurological and clinical biomarker evidence that is collected through use of ripple (80–250 Hz) and fast ripple (250–600 Hz) bands.
In a clinical situation, the number of SOZ and non-SOZ channels in the cortex area depends on the patient, which creates an imbalanced problem. In our survey, only two recent studies related to the design of a computer-aided solution, those proposed by Akter et al. (Akter et al. 2020a, b) addressed these unresolved questions. In their first study, the information-theoretic entropy features were investigated in order to design a computer-aided solution for identification of the SOZ channels of patients with drug-resistant epilepsy. The same research team recently observed the statistical features of high-frequency bands of interictal iEEG that efficiently identify the epileptic focus channels. To assess the possibility of building a patient-independent design (PID), they compared their proposed patient-dependent system with a patient-independent system for localizing SOZ channels through use of the optimal classifier LGBM. Ultimately, their system to extract features from high-frequency iEEG signals and thereby detect epileptic focus completely satisfies neurological hospital measurement standards and complements recent research findings related to epileptic seizure focus. However, many investigations are still required to improve the system’s performance and usability (Akter et al. 2020b).
Neural networks: end-to-end approach
With the rapid growth of neural-network-based methodologies in recent decades, the computer-aided designs that use these methods have been introduced to assist clinical experts in the diagnosis and treatment of epilepsy.
Structure of neural network
The most important aspects of a neural network in this context are the convolutional layer, recurrent layer, fully connected layer, pooling layer, activation function, and batch normalization, among others.
-
The convolutional layer consists of a set of learnable filters (or kernels), each of which has a small receptive field. Dot product (inner product) is performed between the filter weights and region in the input data. The output of the convolutional layer is called the feature map; its depth can be controlled by the number of filters. The stride is set to control how much the filter convolves across the input data.
-
The recurrent layer operates within the cyclical nature of data input and output; each output builds upon the one before it. The RNN with a tanh activation function can be defined as:
$$\begin{aligned} h_{t} = \tanh (W_{x}x_{t} + W_{h}h_{t-1} + b), \end{aligned}$$(4)where \(x_{t}\) is the input data of the time t, and \(h_{t}\) and \(h_{t-1}\) are the hidden states of the time t and \(t-1\), respectively. \(W_{x}\) and \(W_{h}\) are the learnable parameter matrices used for learning input data and hidden state, respectively. b is the parameter vector of the bias.
-
The pooling layer performs the downsampling operation for the input data, which can lower the calculation complexity and prevent overfitting. Some commonly-used pooling operations are max pooling and average pooling, both of which partition the input data into a subregion set. For each subregion, the output is either the maximum or the average value.
-
The fully connected layer is used to compute the class scores in the last layer. A one-dimensional feature vector immediately precedes this layer and functions as its input. In a fully-connected layer, each neuron is connected to all the numbers in the previous volume, which is identical to the traditional multi-layer perceptron neural network.
-
The activation function is a non-linear mathematical operation between the current neuron and its output to the next layer. The definitions of some of the commonly-used activation functions are as follows: \(\text{ Sigmoid } (x) = \frac{1}{1 + e^{-x}}\), \(\tanh (x) = \frac{e^{x} - e^{-x}}{e^{x} + e^{-x}}\), and \(\text{ ReLU }(x) = \max (0, x)\), in each of these, x is the input variable.
-
The batch normalization layer is used for re-centering and re-scaling the input data, which stabilizes the neural networks by allowing for faster convergence. The calculation includes two steps. The first calculates the mean \(E(\cdot )\) and variance \(\text{ Var }(\cdot )\) of a batch data. In the second step, each sample is centered by subtracting the mean and dividing it by the standard deviation: \(y = \frac{x - E(x)}{\sqrt{\text{ Var }(x)}}\), where x is the input variable and y is the normalized result.
Two typical neural network models (CNN and RNN models) are often used to process epilepsy brain signals. The CNN model can capture the waveform features of the EEG signals through the convolution operation. As one of the most widely-used convolutional neural network is VGG model (Simonyan and Zisserman 2015), the CNN achieved first and second places in the localization and classification tracks, respectively, in the Large Scale Visual Recognition Challenge (ILSVRC) of 2014. The model structure is shown in Table 6. Due to the effectiveness of this model in the field of machine-learning approaches, several studies have recently proposed a variant of one-dimensional convolution models for epilepsy signal processing.
Unlike the other neural networks, the RNN uses an internal memory unit to process the result of the previous neural unit. However, the brain signals are time sequence data that depend upon causal relationships with the data before and after. Therefore, the RNN model is suitable for processing brain signals. A basic unit of the RNN model is shown in Fig 6.
Methods based on neural networks
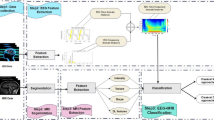
In recent years, neural network methods have been applied to assist with diagnosis and treatment of epilepsy. The use of neural networks is mainly divided into two types. In the first type, feature extraction is performed through an algorithm, and then the neural network is used as a classifier. In the second type, feature extraction and classifier are all done by the neural network in an end-to-end model. Two commonly-used neural network models are show in Fig. 7.
Sui et al. (2019) used short-time Fourier transform (STFT) method to extract time-frequency features of the EEG signals. The obtained features were used as an input to the CNN model. Subathra et al. (2020) used the fast Walsh-Hadamard transform (FWHT) method to analyze the EEG signals in the frequency domain and decomposed them into the Hadamard coefficients. Five different nonlinear features were extracted from the decomposed Hadamard coefficients: approximate entropy (ApEn), log-energy entropy (LogEn), fuzzy entropy (FuzzyEn), sample entropy (SampEn), and permutation entropy (PermEn).To discriminate focal and non-focal channels, the artificial neural network (ANN) was used as a classifier. Siddharth et al. (2019) decomposed the EEG signals by using sliding mode-singular spectrum analysis (SM-SSA). Next, the classifier was designed by combining the sparse-autoencoder (SAE) hidden layer and the radial basis function neural network (RBFN). By using multiple filters and entropy (Zhao et al. 2018), features matrices were extracted from EEG signals, then focal and non-focal features were classified based on CNN method. San-Segundo et al. (2019)proposed a convolutional and fully-connected layers as classifier using a variant of multi-band decomposition analysis such as FT, WT, and EMD. Gagliano et al. (2019) introduced bispectrum analysis as the signal features; it resulted in a two-dimensional map** of nonlinear interactions between the various frequency components of a time series. Last, the classification was implemented by using long-short-term memory (LSTM). Recently, by taking into account the feature interaction information, Zhao et al. (2021) integrated the tensor fusion strategy into deep CNN method for identifying epileptic focus.
The end-to-end method refers to training a possibly complex learning system represented by a model and avoiding the traditional feature process. Daoud and Bayoumi (2019) automated and integrated extracted features and the classification processes by using both a deep convolutional auto-encoder and multi-layer perceptron. Due to the EEG signal being one-dimensional data, Li et al. (2019) used a one-dimensional convolutional neural network model to perform the feature extraction and classification functions at the same time. In a recent epilepsy machine-learning study, Lu and Triesch (2019) exploited an automated epileptic signal classification method based on deep neural networks with residual connections. To observe the efficacy of their proposed method, two datasets, including Bonn University and the Bern-Barcelona data sets, were used for two kind of separate classification problems in epilepsy, such as the epileptic seizure pattern identification from continuous EEG signals (Bonn University sets) and localization of epileptic focus (Bern-Barcelona data sets). Fraiwan and Alkhodari (2020) used an LSTM model to classify the focal and non-focal epileptic EEG data directly from input. Jukic et al. (2020)compared the effects of the ensemble learning method with different machine learning models for the localization of epileptic focus. Table 7 gives a summary of the previously-described neural network methods.
Evaluation criteria
The evaluation criteria are the key factors that measure the efficacy of the computer-aided solutions. For completeness, a brief review of the concepts of evaluation of performance are presented to know what matrices must be considered during evaluation computer-aided solutions to the problem of epileptic focus localization.
Segment-wise criteria
A long-term EEG recording is usually segmented into short-time epochs to classify focal and non-focal channels. Therefore, for each segment (epoch), typical metrics can be used to evaluate performance. This section summarizes the evaluation metrics used in related works to identify epileptic seizure focus. The most basic performance evaluation metrics used in different epileptic focus detection studies are accuracy, sensitivity, specificity, precision, false discovery rate (FDR), and \(F_1\)-scores. However, classification accuracy is valid only if the classes are balanced (Fatourechi et al. 2008; Dornhege et al. 2004), which has the same performance for each class. For imbalanced classes, the sensitivity (SEN), specificity (SPE), precision, and fall-out or false-negative rate (FPR) are more informative performance measures (Fatourechi et al. 2008; Dornhege et al. 2004), and can be computed from the confusion matrix. If the classification depends on a continuous parameter, such as threshold, the receiver operating characteristic (ROC) curve and the area under the curve (AUC) are often used. The representation of classification performance can be computed from the confusion matrix, as illustrated in Table 8. Based on that table, the evaluation metrics can be defined as:
-
Accuracy (ACC):
$$\begin{aligned} Accuracy=\frac{TP+TN}{TP+FP+TN+FN}\times 100\%, \end{aligned}$$(5) -
Sensitivity (SEN) or recall:
$$\begin{aligned} SEN=\frac{TP}{TP+FN}\times 100, \end{aligned}$$(6) -
Specificity (SPE):
$$\begin{aligned} SPE=\frac{TN}{TN+FP}\times 100, \end{aligned}$$(7) -
Precision or postitive predictive value (PPV):
$$\begin{aligned} Precision=\frac{TP}{TP+FP}\times 100, \end{aligned}$$(8) -
Fall-out or false positive rate (FPR):
$$\begin{aligned} FPR_{nfocal}=\frac{FP}{TN+FP}\times 100, \end{aligned}$$(9) -
and F\(_{1}\) score, which is the harmonic mean of preision and sensitivity defined as:
$$\begin{aligned} F_{1}-score=\frac{2}{\frac{1}{Recall}+\frac{1}{precision}}, \end{aligned}$$(10)
where TP is the number of correctly-detected focal segments, and FN indicates the number of incorrectly-detected focal segments. TN is the number of correctly-detected non-focal segments, and FP represents the number of incorrectly-detected non-focal segments.
Electrode-wise evaluation criteria
Recently, Akter et al. (2020a, 2020b) introduced a score-based evaluation approach in each electrode by counting correctly-detected focal segments, which can be considered as a more effective evaluation criteria for detecting epileptic focus. In conventional clinical systems, epileptologists need to observe long-term multi-channel iEEG signals by dividing them into multiple segments (in their case, 20 s for each segment) to find the epileptic focus. The performance estimation from individual segments does not provide appropriate statistic. Thus, Akter et al. (2020a, 2020b) scored each channel with a number by the correctly-detected focal segments. The SEN and false positive rate (FPR) were estimated for each threshold value from the channel scores. After achieving SEN and PFR with each threshold value, the AUC under receiver operating curve (ROC) was estimated using the trapezoid rule (Fawcett 2006). An example of performance measurement with channels is shown in Fig. 8. It shows an epileptic focus-diagnostic illustration, which can help the epileptologists in two ways: (i) by facilitating observation of the localization of the focal and non-focal segments throughout the multi-channel iEEG signals (as seen on the left side of the Fig. 8). Each yellow spot in the color map represents the detected focal segments; (ii) the number of detected focal segments corresponding to the seizure and non-seizure onset channels (right side of Fig. 8). Figure 8 indicates the number of detected focal segments (x-axis) in each electrode (y-axis) in which the red bars represent the SOZ and the black bars indicates a non-SOZ area. The AUC result for identifying SOZ and non-SOZ channels was estimated for each threshod value from the channel scores shown in right side of Fig. 8. The reader is referred to the work by Akter et al. (2020a, 2020b) on evaluation of computer-aided design for identification of SOZ channels.
The colormap representation on the left-hand side illustrates the channels (y-axis) and segment index (x-axis). Each yellow spot in the color map indicates detected focal segments. The right side of the color map (bar) represents the number of detected focal segments (x-axis) in each electrode (y-axis). The red bars indicate the SOZ channels labeled by epileptologists
Discussion and open problems
This survey aimed to provide a comprehensive overview of the current epilepsy research focused on develo** a computer-aided system for the localization of epileptic focus detection. We have surveyed the different biomarkers- and feature-extraction-based approaches developed between 2013 and 2020 to facilitate creation of an AI-based diagnosis-aid. The numerous explored approaches can be divided into four main categories: (i) HFOs, (ii) PAC, (iii) IED, and (iv) feature-extraction methods.
Some epilepsy studies with a variant of public datasets, including Bonn, Flint Hill, Freiburg, CHB-MIT, Epilepsia, and TUSZ, introduced computer-aided solutions to identify epilepsy patients or seizure patterns in EEG signals. For instance, the Bonn dataset consists of five subjects with single-channel surface EEG at a sampling rate of 173.61 Hz, denoted as Set A (healthy awake and eyes open), Set B (healthy awake and eyes closed), Set C (Epileptic seizure-free interictal), Set D (Epileptic seizure-free interictal), and Set E (epileptic seizure activity). A two-class problem was formed by assigning class labels to epileptic activity (set A) vs others (Set B, C, D, and E). Therefore, different combinations, such as E vs. A, E vs. B, E vs. C, E vs. D, and so on, were performed to detect epileptic vs. non-epileptic activities. However, this dataset’s main goal was to detect epilepsy patients from other subjects. Similarly, the other datasets listed in Table 2 only focused on detecting either seizure or non-seizure patterns in an EEG segment.
For drug-resistant epilepsy, the primary clinical problem is identifying the epileptic focus that must be removed entirely in order to halt seizures. To identify the epileptic focus for surgery, recent neurology hospitals have used grid electrodes (multi-channel) with a high sampling rate to observe ripple (80–250 Hz) and fast ripple (250–600 Hz) bands. The underlying idea behind using a high sampling rate is that ripple and fast ripple bands bear the potential guide for surgical treatment of drug-resistant focal epilepsy. Of note, the epilepsy biomarkers (HFOs, PAC, and IEDs) may occur during ictal, preictal, and interictal states, and the rate of biomarker events tends to be higher in the SOZ channels. As discussed in the biomarker section, many research works based on AI aim to develop automated methods for finding these biomarkers in multi-channel EEG signals. However, the biomarkers in EEGs are differentiated into ripples and fast ripples in EEG signals, and the identification of the biomarkers requires separate ripple and fast ripple iEEGs to identify SOZ channels. When designing a computer-aided solution using these biomarkers, most of the algorithms encounter a common problem: precisely calculating baseline activity. This is because the background activity is not perfectly flat in multi-channel EEG. It is notable that there are several possible subgroups of neurological biomarkers in EEG signals, including HFOs (ripples, fast ripples, and ripples+fast ripples) and IEDs (spikes, wave complex, polyspike, slow-wave, and artifacts). These biomarkers’ variants collectively hint at an intricate AI solution for the identification of possible SOZ channels. However, due to the lack of a standard public dataset related to the biomarkers and their distinctive performance evaluation, it is infeasible to compare the literature’s biomarker-related AI systems.
According to the various studies in this paper, many techniques have introduced the feature-extraction methods described in Table 5 to identify epileptic focus from EEG signals collected in public and hospital datasets. The studies listed in Table 5 collectively provide evidence that the information-theoretic and statistical features are another engineering marker that may help researchers to identify the epileptic focus. Among the many significant advantages of designing a computer-aided system is that it does not need to find a variant of biomarkers in EEG signals, as the statistical features measure the data dispersion from EEG signals to characterize normal and epileptic activities. It is important to note that EEG signals recorded from epilepsy patients may consist of epileptic biomarkers during ictal and interictal phases. The use of statistical methods may provide exciting tools that allow scholars to isolate the normal and epileptic properties from EEG signals and eventually design computer-aided solutions for the identification of SOZ channels.
Based on our survey, we also wish to identify some research problems that must be resolved before more efficient computer-aided solutions can be designed following:
-
In our survey, recent studies that utilize engineering solutions to identify SOZ channels have shown promising results. Different ages and pathological types with an increased number of patients should be considered for future studies.
-
Most of the studies in this survey focused on develo** patient-dependent methods to improve computer-aided systems, not on a patient-independent system (PID). For real-world applications, indeed, the patient-independent design (PID) is preferable because epileptologists require some EEG data to label focal and non-focal electrodes used in the system to hypothesis the possible SOZ channels. However, the design of a patient-independent system for identifying SOZ channels is challenging due to the very different electrodes and subject-specific nature of EEG signals. The most promising directions, those that allow for adaptation to different distributions, could be transfer learning and domain adaptation (Pan and Yang 2009; Lotte and Guan 2010; Azab et al. 2018).
-
For designing a supervised computer-aided system (either patient-dependent or patient-independent), the major limitation is the necessity to use SOZ as a prior-basis ground truth for the classifier training stage. Therefore, designing an unsupervised computer-aided system for identification of the SOZ can provide a great facility without prior-basis information of ground truth to take a medical decision.
-
Data recording for clinical protocol depends on the patient’s conditions. In particular, it is challenging to collect enough data to apply to machine learning. Data augmentation is another hot topic in AI design, as it could be used to improve system usability and reduce the training set. Some attempts at data augmentation of focus detection have been reported (Akter et al. 2020a, b).
-
The selection of influential parameters is another critical factor to the design of a computer-aided system. Parameters with more intelligent signal processing and feature-extraction methods are required to further improve focus identification performance.
-
Statistical and information-theoretic features in high-frequency components are promising in this application; however, the interpretation of these features in terms of clinical neurophysiology are still in question.
To conclude, this review suggests that the development of robust, efficient, and convenient computer-aided systems based on feature-extraction methods for identifying SOZ channels provides a new direction in epilepsy research. The development of a trustworthy system remains an open challenge. The next generation of machine-learning approaches to computer-aided diagnosis systems will have to take the user into account. Furthermore, we would like to emphasize that the collaboration between engineering and medical sectors is the most crucial key to the success of a practical computer-aided diagnosis aid.
References
Abd El-Samie FE, Alotaiby TN, Khalid MI, Alshebeili SA, Aldosari SA (2018) A review of EEG and MEG epileptic spike detection algorithms. IEEE Access 6:60673–60688
Acharya UR, Hagiwara Y, Deshpande SN, Suren S, Koh JEW, Oh SL, Arunkumar N, Ciaccio EJ, Lim CM (2019) Characterization of focal EEG signals: a review. Future Gener Comput Syst 91:290–299
Acır N, Güzeliş C (2004) Automatic spike detection in EEG by a two-stage procedure based on support vector machines. Comput Biol Med 34(7):561–575
Adjouadi M, Cabrerizo M, Ayala M, Sanchez D, Yaylali I, Jayakar P, Barreto A (2004) A new mathematical approach based on orthogonal operators for the detection of interictal spikes in epileptogenic data. Biomed Sci Instrum 40:175–180
Adjouadi M, Sanchez D, Cabrerizo M, Ayala M, Jayakar P, Yaylali I, Barreto A (2004) Interictal spike detection using the Walsh transform. IEEE Trans Biomed Eng 51(5):868–872
Akter MS, Islam MR, Iimura Y, Sugano H, Fukumori K, Wang D, Tanaka T, Cichocki A (2020) Multiband entropy-based feature-extraction method for automatic identification of epileptic focus based on high-frequency components in interictal iEEG. Sci Rep 10(1):1–17
Akter MS, Islam MR, Tanaka T, Iimura Y, Mitsuhashi T, Sugano H, Wang D, Molla MKI (2020) Statistical features in high-frequency bands of interictal iEEG work efficiently in identifying the seizure onset zone in patients with focal epilepsy. Entropy 22(12):1415
Akter MSA, Islam MRI, Tanaka TT, Fukumori KF, Iimura YI, Sugano HS (2019) Automatic identification of epileptic focus on high–frequency components in interictal iEEG. In: 2019 8th International congress on advanced applied informatics (IIAI–AAI), pp 1075–1076
Amiri M, Frauscher B, Gotman J (2016) Phase-amplitude coupling is elevated in deep sleep and in the onset zone of focal epileptic seizures. Front Human Neurosci 10:387
Amiri M, Frauscher B, Gotman J (2019) Interictal coupling of HFOs and slow oscillations predicts the seizure-onset pattern in mesiotemporal lobe epilepsy. Epilepsia 60:1160–1170
Andrzejak RG, Lehnertz K, Mormann F, Rieke C, David P, Elger CE (2001) Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: dependence on recording region and brain state. Phys Rev E 64(6):061907
Andrzejak RG, Schindler K, Rummel C (2012) Nonrandomness, nonlinear dependence, and nonstationarity of electroencephalographic recordings from epilepsy patients. Phys Rev E 86(4):046206
Antony AR, Alexopoulos AV, González-Martínez JA, Mosher JC, Jehi L, Burgess RC, So NK, Galán RF (2013) Functional connectivity estimated from intracranial eeg predicts surgical outcome in intractable temporal lobe epilepsy. PloS One 8(10):e77916
Arunkumar A, Ramkumar RK, Venkatraman VV, Abdulhay E, Fernandes SL, Kadry S, Segal S (2017) Classification of focal and non focal eeg using entropies. Pattern Recognit Lett 94:112–117
Azab AM, Toth J, Mihaylova LS, Arvaneh M (2018) Signal processing and machine learning for brain-machine interfaces. IET London, UK
Azami H, Sanei S (2014) Spike detection approaches for noisy neuronal data: assessment and comparison. Neurocomputing 133:491–506
Bancaud J, Talairach J (1992) Clinical semiology of frontal lobe seizures. Adv Neurol 57:3–58
Barlow JS (1980) EEG transient detection by matched inverse digital filtering. Electroencephalogr Clin Neurophysiol 48(2):246–248
Bhattacharyya A, Pachori RB, Acharya UR (2017) Tunable-Q wavelet transform based multivariate sub-band fuzzy entropy with application to focal EEG signal analysis. Entropy 19(3):99
Bhattacharyya A, Sharma M, Pachori RB, Sircar P, Acharya UR (2018) A novel approach for automated detection of focal EEG signals using empirical wavelet transform. Neural Comput Appl 29(8):47–57
Birot G, Kachenoura A, Albera L, Bénar C, Wendling F (2013) Automatic detection of fast ripples. J Neurosci Methods 213(2):236–249
Black MA, Jones RD, Carroll GJ, Dingle AA, Donaldson IM, Parkin PJ (2000) Real-time detection of epileptiform activity in the EEG: a blinded clinical trial. Clin EEG Neurosci 31(3):122–130
Blanco JA, Stead M, Krieger A, Viventi J, Marsh WR, Lee KH, Worrell GA, Litt B (2010) Unsupervised classification of high-frequency oscillations in human neocortical epilepsy and control patients. J Neurophysiol 104(5):2900–2912
Boos CF, de Azevedo FM, Scolaro GR, Do Pereira M (2011) Automatic detection of paroxysms in EEG signals using morphological descriptors and artificial neural networks. Biomedical engineering, trends in electronics. Communications and software. InTech, Rijeka, pp 387–402
Bourien J, Bellanger JJ, Bartolomei F, Chauvel P, Wendling F (2004) Mining reproducible activation patterns in epileptic intracerebral EEG signals: application to interictal activity. IEEE Trans Biomed Eng 51(2):304–315
Bénar C, Chauvière L, Bartolomei F, Wendling F (2010) Pitfalls of high-pass filtering for detecting epileptic oscillations: a technical note on false ripples. Clin Neurophysiol 121(3):301–310
Canolty R, Edwards E, Dalal S, Soltani M, Nagarajan S, Kirsch H, Berger M, Barbaro N, Knight R (2006) High gamma power is phase-locked to theta oscillations in human neocortex. Science 313(5793):1626–1628
Chaibi S, Lajnef T, Sakka Z, Samet M, Kachouri A (2013) A comparaison of methods for detection of high frequency oscillations (HFOs) in human intacerberal EEG recordings. Am J Signal Process 3(2):25–34
Chaibi S, Lajnef T, Samet M, Jerbi K, Kachouri A (2014) Detection of high frequency oscillations (hfos) in the 80–500 hz range in epilepsy recordings using decision tree analysis. In: International image processing, applications and systems conference, pp 1–6
Chavakula V, Fernández IS, Peters JM, Popli G, Bosl W, Rakhade S, Rotenberg A, Loddenkemper T (2013) Automated quantification of spikes. Epilepsy Behav 26(2):143–152
Chen D, Wan S, Bao FS (2017) Epileptic focus localization using discrete wavelet transform based on interictal intracranial eeg. IEEE Trans Neural Syst Rehabil Eng 25(5):413–425
Crépon B, Navarro V, Hasboun D, Clemenceau S, Martinerie J, Baulac M, Adam C, Le Van Quyen M (2010) Map** interictal oscillations greater than 200 Hz recorded with intracranial macroelectrodes in human epilepsy. Brain 133(1):33–45
Dalal M, Tanveer M, Pachori RB (2019) Automated identification system for focal EEG signals using fractal dimension of FAWT–based sub–bands signals. In: Machine Intelligence and Signal Analysis, Springer, pp 583–596
Daoud H, Bayoumi M (2019) Deep learning approach for epileptic focus localization. IEEE Trans Biomed Circuits Syst 14(2):209–220
Das AB, Bhuiyan MIH (2016) Discrimination and classification of focal and non-focal EEG signals using entropy-based features in the EMD-DWT domain. Biomed Signal Process Control 29:11–21
De Curtis M, Avanzini G (2001) Interictal spikes in focal epileptogenesis. Prog Neurobiol 63(5):541–567
De Moraes FD, Callegari DA (2014) Automated detection of interictal spikes in EEG: a literature review. Clin Neurophysiol, pp 1095–1103
Deivasigamani S, Senthilpari C, Yong WH (2016) Classification of focal and nonfocal EEG signals using ANFIS classifier for epilepsy detection. Int J Imaging Syst Technol 26(4):277–283
Deuschl G (1999) Recommendations for the practice of clinical neurophysiology: guidelines of the international federation of clinical neurophysiology. Electroencephalogr Clin Neurophysiol Suppl 52:1–304
Dornhege G, Blankertz B, Curio G, Müller KR (2004) Boosting bit rates in noninvasive EEG single-trial classifications by feature combination and multiclass paradigms. IEEE Trans Biomed Eng 51(6):993–1002
Durka PJ (2004) Adaptive time-frequency parametrization of epileptic spikes. Phys Rev E 69(5):051914
El-Gohary M, McNames J, Elsas S (2008) User–guided interictal spike detection. In: 2008 30th annual International conference of the IEEE engineering in medicine and biology society (EMBC), pp 821–824
Elahian B, Yeasin M, Mudigoudar B, Wheless JW, Babajani-Feremi A (2017) Identifying seizure onset zone from electrocorticographic recordings: a machine learning approach based on phase locking value. Seizure 51:35–42
Elsharkawy AE, Pietilä TA, Alabbasi AH, Pannek H, Ebner A (2011) Long term outcome in patients not initially seizure free after resective epilepsy surgery. Seizure 20(5):419–424
Exarchos TP, Tzallas AT, Fotiadis DI, Konitsiotis S, Giannopoulos S (2006) EEG transient event detection and classification using association rules. IEEE Trans Inf Technol Biomed 10(3):451–457
Fatourechi M, Ward RK, Mason SG, Huggins J, Schlögl A, Birch GE (2008) Comparison of evaluation metrics in classification applications with imbalanced datasets. In: 2008 seventh International conference on machine learning and applications, pp 777–782
Fawcett T (2006) An introduction to roc analysis. Pattern Recognit Lett 27(8):861–874
Fedele T, van’t Klooster M, Burnos S, Zweiphenning W, van Klink N, Leijten F, Zijlmans M, Sarnthein J (2016) Automatic detection of high frequency oscillations during epilepsy surgery predicts seizure outcome. Clin Neurophysiol 127(9):3066–3074
Fedele T, Ramantani G, Sarnthein J (2019) High frequency oscillations as markers of epileptogenic tissue-end of the party? Clin Neurophysiol Off J Int Fed Clin Neurophysiol 130(5):624–626
Feucht M, Hoffmann K, Steinberger K, Witte H, Benninger F, Arnold M, Doering A (1997) Simultaneous spike detection and topographic classification in pediatric surface EEGs. NeuroReport 8(9):2193–2197
Firpi H, Smart O, Worrell G, Marsh E, Dlugos D, Litt B (2007) High-frequency oscillations detected in epileptic networks using swarmed neural-network features. Ann Biomed Eng 35(9):1573–1584
Fischer G, Mars N, da Silva FL (1980) Pattern recognition of epileptiform transients in the electroencephalogram. Institute of Medical Physics, Utrecht
Fisher RS, Acevedo C, Arzimanoglou A, Bogacz A, Cross JH, Elger CE, Engel J, Forsgren L, French JA, Glynn M, Hesdorffer DC, Lee B, Mathern GW, Moshé SL, Perucca E, Scheffer IE, Tomson T, Watanabe M, Wiebe S (2014) ILAE official report: a practical clinical definition of epilepsy. Epilepsia 55(4):475–482
Fraiwan L, Alkhodari M (2020) Classification of focal and non-focal epileptic patients using single channel EEG and long short-term memory learning system. IEEE Access 8:77255–77262
Fukumori K, Nguyen HTT, Yoshida N, Tanaka T (2019) Fully data–driven convolutional filters with deep learning models for epileptic spike detection. In: 2019-2019 International conference on acoustics, speech and signal processing (ICASSP), pp 2772–2776
Fürbass F, Hartmann M, Halford J, Koren J, Herta J, Gruber A, Baumgartner C, Kluge T (2015) Automatic detection of rhythmic and periodic patterns in critical care EEG based on American Clinical Neurophysiology Society (ACNS) standardized terminology. Clin Neurophysiol 45(3):203–213
Gagliano L, Assi EB, Nguyen DK, Sawan M (2019) Bispectrum and recurrent neural networks: improved classification of interictal and preictal states. Sci Rep 9(1):1–9
Gardner AB, Worrell GA, Marsh E, Dlugos D, Litt B (2007) Human and automated detection of high-frequency oscillations in clinical intracranial EEG recordings. Clin Neurophysiol 118(5):1134–1143
Garg HK, Kohli AK (2013) Nonstationary-epileptic-spike detection algorithm in EEG signal using sneo. Biomed Eng Lett 3(2):80–86
Giannakakis G, Sakkalis V, Pediaditis M, Tsiknakis M (2014) Methods for seizure detection and prediction: an overview. Neuromethods 91:131–157
Gliske SV, Irwin ZT, Davis KA, Sahaya K, Chestek C, Stacey WC (2016) Universal automated high frequency oscillation detector for real-time, long term EEG. Clin Neurophysiol 127(2):1057–1066
Glover JR, Raghaven N, Ktonas PY, Frost JD (1989) Context-based automated detection of epileptogenic sharp transients in the EEG: elimination of false positives. IEEE Trans Biomed Eng 36(5):519–527
Goelz H, Jones RD, Bones PJ (2000) Wavelet analysis of transient biomedical signals and its application to detection of epileptiform activity in the EEG. Clin Electroencephalogr 31(4):181–191
Goldberger A, Amaral L, Glass L, Hausdorff J, Ivanov PC, Mark R, Mietus J, Moody G, Peng C, Stanley H (2000) PhysioBank, PhysioToolkit, and Physionet. Circulation 101(23):e215–e220
Gopan KG, Harsha A, Joseph LA, Kollialil ES (2013) Adaptive neuro–fuzzy classifier for ‘petit mal’ epilepsy detection using mean teager energy. In: 2013 International conference on advances in computing, communications and informatics (ICACCI), pp 752–757
Gotman J, Gloor P (1976) Automatic recognition and quantification of interictal epileptic activity in the human scalp EEG. Electroencephalogr Clin Neurophysiol 41(5):513–529
Guirgis M, Chinvarun Y, Campo MD, Carlen P, Bardakjian B (2015) Defining regions of interest using cross-frequency coupling in extratemporal lobe epilepsy patients. J Neural Eng 12(2):026011
Gupta V, Pachori RB (2020) Classification of focal EEG signals using FBSE based flexible time-frequency coverage wavelet transform. Biomed Signal Process Control 62:102–124
Gupta V, Priya T, Yadav AK, Pachori RB, Rajendra Acharya U (2017) Automated detection of focal EEG signals using features extracted from flexible analytic wavelet transform. Pattern Recognit Lett 94:180–188
Halford JJ (2009) Computerized epileptiform transient detection in the scalp electroencephalogram: obstacles to progress and the example of computerized ECG interpretation. Clin Neurophysiol 120(11):1909–1915
Halford JJ, Schalkoff RJ, Zhou J et al (2013) Standardized database development for EEG epileptiform transient detection: Eegnet scoring system and machine learning analysis. J Neurosci Methods 212(2):308–316
Harati A, Lopez S, Obeid I, Picone J, Jacobson M, Tobochnik S (2014) The TUH EEG CORPUS: A big data resource for automated EEG interpretation. In: 2014 IEEE signal processing in medicine and biology symposium (SPMB), pp 1–5
Hassan KM, Islam MR, Tanaka T, Molla MKI (2019) Epileptic seizure detection from EEG signals using multiband features with feedforward neural network. In: 2019 International conference on cyberworlds (CW), pp 231–238
Herta J, Koren J, Fürbass F, Hartmann M, Kluge T, Baumgartner C, Gruber A (2015) Prospective assessment and validation of rhythmic and periodic pattern detection in neurotrend: a new approach for screening continuous EEG in the intensive care unit. Epilepsy Behav 49:273–279
Hese PV, Vanrumste B, Hallez H, Carroll GJ, Vonck K, Jones RD, Bones PJ, D’Asseler Y, Lemahieu I (2008) Detection of focal epileptiform events in the EEG by spatio-temporal dipole clustering. Clin Neurophysiol 119(8):1756–1770
Hussein AF, Arunkumar N, Gomes C, Alzubaidi AK, Habash QA, Santamaria-Granados L, Mendoza-Moreno JF, Ramirez-Gonzalez G (2018) Focal and non-focal epilepsy localization: a review. IEEE Access 6:49306–49324
Ihle M, Feldwisch-Drentrup H, Teixeira CA, Witon A, Schelter B, Timmer J, Schulze-Bonhage A (2012) EPILEPSIAE-A European epilepsy database. Comput Methods Programs Biomed 106(3):127–138
Isaksson A, Wennberg A, Zetterberg LH (1981) Computer analysis of EEG signals with parametric models. Proc IEEE 69(4):451–461
Islam MK (2015) Artifact characterization, detection and removal from neural signals. PhD thesis, National University of Singapore, Singapore
Islam MR, Tanaka T, Molla MKI (2018) Multiband tangent space map** and feature selection for classification of EEG during motor imagery. J Neural Eng 15(4):046021
Itakura T, Tanaka T (2017) Epileptic focus localization based on bivariate empirical mode decomposition and entropy. In: Asia–Pacific signal and information processing association annual summit and conference (APSIPA ASC), pp 1426–1429
Jacobs J, Zelmann R, Jirsch J, Chander R, Dubeau CCF, Gotman J (2009) High frequency oscillations (80–500 hz) in the preictal period in patients with focal seizures. Epilepsia 50(7):1780–1792
Jacobs J, Wu JY, Perucca P, Zelmann R, Mader M, Dubeau F, Mathern GW, Schulze-Bonhage A, Gotman J (2018) Removing high-frequency oscillations: a prospective multicenter study on seizure outcome. Neurology 91(11):e1040–e1052
Ji Z, Sugi T, Goto S, Wang X, Ikeda A, Nagamine T, Shibasaki H, Nakamura M (2011) An automatic spike detection system based on elimination of false positives using the large-area context in the scalp EEG. IEEE Trans Biomed Eng 58(9):2478–2488
Ji Z, Sugi T, Goto S, Wang X, Nakamura M (2011b) Multi–channel template extraction for automatic EEG spike detection. In: IEEE/ICME International conference on complex medical engineering, pp 179–184
Jrad N, Kachenoura A, Merlet I, Nica A, Benar CG, Wendling F (2015) Classification of high frequency oscillations in epileptic intracerebral EEG. In: Proceedings of the annual international conference of the IEEE engineering in medicine and biology society, (EMBS), pp 574–577
Jrad N, Kachenoura A, Merlet I, Bartolomei F, Nica A, Biraben A, Wendling F (2017) Automatic detection and lassification of high-frequency oscillations in depth-EEG signals. IEEE Trans Biomed Eng 64(9):2230–2240
Jukic S, Saracevic M, Subasi A, Kevric J (2020) Comparison of ensemble machine learning methods for automated classification of focal and non-focal epileptic EEG signals. Mathematics 8(9):1481
Juozapavicius A, Bacevicius G, Bugelskis D, Samaitiene R (2011) EEG analysis-Automatic spike detection. Nonlinear Anal Model Control 16(4):375–386
Kalayci T, Ozdamar O (1995) Wavelet preprocessing for automated neural network detection of EEG spikes. IEEE Eng Med Biol Mag 14(2):160–166
Khalilov I, Le Van Quyen M, Gozlan H, Ben-Ari Y (2005) Epileptogenic actions of GABA and fast oscillations in the develo** hippocampus. Neuron 48(5):787–796
Klimes P, Duque JJ, Jurak P, Halamek J, Worrell GA (2015) Connectivity of epileptic brain regions in wake and sleep. In: proceedings of the annual International conference of the IEEE engineering in medicine and biology society, (EMBC), pp 2191–2194
Koren J, Herta J, Draschtak S, Pötzl G, Pirker S, Fürbass F, Hartmann M, Kluge T, Baumgartner C (2015) Prediction of rhythmic and periodic EEG patterns and seizures on continuous EEG with early epileptiform discharges. Epilepsy Behav 49:286–289
Levesque M, Shiri Z, Chen L, Avoli M (2017) High-frequency oscillations and mesial temporal lobe epilepsy. Neurosci Lett 667:66–74
Li B, Zhao X, Zhao Q, Tanaka T, Cao J (2019) A one–dimensional convolutional neural network model for automated localization of epileptic foci. In: Asia–Pacific signal and information processing association annual summit and conference (APSIPA ASC), pp 741–744
Liu S, Sha Z, Sencer A, Aydoseli A, Bebek N, Abosch A, Henry T, Gurses C, Ince NF (2016) Exploring the time-frequency content of high frequency oscillations for automated identification of seizure onset zone in epilepsy. J Neural Eng 13(2):026026
Liu YC, Lin CCK, Tsai JJ, Sun YN (2013) Model-based spike detection of epileptic EEG data. Sensors 13(9):12536–12547
Lodder SS, Askamp J, van Putten MJ (2013) Inter-ictal spike detection using a database of smart templates. Clin Neurophysiol 124(12):2328–2335
Logesparan L, Rodriguez-Villegas E (2011) A novel phase congruency based algorithm for online data reduction in ambulatory EEG systems. IEEE Trans Biomed Eng 58(10):2825–2834
Lotte F, Guan C (2010) Learning from other subjects helps reducing brain-computer interface calibration time. In: 2010 IEEE International conference on acoustics, speech and signal processing (ICASSP), pp 614–617
Lu D, Triesch J (2019) Residual deep convolutional neural network for EEG signal classification in epilepsy. ar**v:1903.08100 [math.NA]
Lüders H, Najm I, Nair D, Widdess-Walsh P, Bingman W (2006) The epileptogenic zone: general principles. Epileptic Disorders 8(2):1–9
Matsumoto A, Brinkmann BH, Stead SM, Matsumoto J, Kucewicz MT, Marsh WR, Meyer F, Worrell G (2013) Pathological and physiological high-frequency oscillations in focal human epilepsy. J Neurophysiol 110(8):1958–1964
Medvedev A, Agoureeva G, Murro A (2019) A long short-term memory neural network for the detection of epileptiform spikes and high frequency oscillations. Sci Rep 9(1):1–10
van Mierlo P, Papadopoulou M, Carrette E, Boon P, Vandenberghe S, Vonck K, Marinazzo D (2014) Functional brain connectivity from EEG in epilepsy: seizure prediction and epileptogenic focus localization. Prog Neurobiol 121:19–35
Motoi H, Miyakoshi M, Abel T, Jeong J, Nakai Y, Sugiura A, Luat A, Agarwal R, Sood S, Asano E (2018) Phase-amplitude coupling between interictal high-frequency activity and slow waves in epilepsy surgery. Epilepsia 59:1954–1965
Nakamura M, Ikeda A, Sugi T, Shibasaki H (2001) Adaptive EEG spike detection: determination of threshold values based on conditional probability. Front Med Biol Eng 11(4):261–277
Nariai H, Matsuzaki M, Juhasz C, Nagasawa T, Sood S, Chugani H, Asano E (2011) Ictal high-frequency oscillations at 80–200 Hz coupled with delta phase in epileptic spasms. Epilepsia 52(10):e130–e134
Navarrete M, Alvarado-Rojas C, Le Van Quyen M, Valderrama M (2016) RIPPLELAB: a comprehensive application for the detection, analysis and classification of high frequency oscillations in electroencephalographic signals. PLOS ONE 11(6):1–27
Navarrete M, Pyrzowski J, Corlier J, Valderrama M, Le Van Quyen M (2016) Automated detection of high-frequency oscillations in electrophysiological signals: methodological advances. J Physiol-Paris 110(4, Part A):316–326
Ngugi A, Kariuki S, Bottomley C, Kleinschmidt I, Sander J, Newton C (2011) Incidence of epilepsy. Neurology 77(10):1005–1012
Noachtar S, Binnie C, Ebersole J, Mauguiere F, Sakamoto A, Westmoreland B (1999) A glossary of terms most commonly used by clinical electroencephalographers and proposal for the report form for the EEG findings. The International Federation of Clinical Neurophysiology. Electroencephalogr Clin Neurophysiol Suppl 52:21–41
Nonclercq A, Foulon M, Verheulpen D, De Cock C, Buzatu M, Mathys P, Van Bogaert P (2012) Cluster-based spike detection algorithm adapts to interpatient and intrapatient variation in spike morphology. J Neurosci Methods 210(2):259–265
Obeid I, Picone J (2016) The Temple University Hospital EEG data corpus. Front Neurosci 10:196
Oikonomou VP, Tzallas AT, Fotiadis DI (2007) A Kalman filter based methodology for EEG spike enhancement. Comput Methods Programs Biomed 85(2):101–108
Osorio I, Harrison MAF, Lai YC, Frei MG (2001) Observations on the application of the correlation dimension and correlation integral to the prediction of seizures. J Clin Neurophysiol 18(3):269–274
Özdamar Ö, Kalayci T (1998) Detection of spikes with artificial neural networks using raw EEG. Comput Biomed Res 31(2):122–142
Pan SJ, Yang Q (2009) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359
Pardey J, Roberts S, Tarassenko L (1996) A review of parametric modelling techniques for EEG analysis. Med Eng Phys 18(1):2–11
Pati S, Alexopoulos A (2010) Pharmacoresistant epilepsy: from pathogenesis to current and emerging therapies. Clevel Clin J Med 77:457–467
Paul Y (2018) Various epileptic seizure detection techniques using biomedical signals: a review. Brain Inform 5:6
Pietilä T, Vapaakoski S, Nousiainen U, Värri A, Frey H, Häkkinen V, Neuvo Y (1994) Evaluation of a computerized system for recognition of epileptic activity during long-term EEG recording. Electroencephalogr Clin Neurophysiol 90(6):438–443
Radmehr M, Anisheh SM (2013) EEG spike detection using stationary wavelet transform and time–varying autoregressive model. Int J Comput Appl 83(13)
Rilling G, Flandrin P, Gonçalves P, Lilly JM (2007) Bivariate empirical mode decomposition. IEEE Signal Process Lett 14(12):936–939
Rosenow F, Lüders H (2001) Presurgical evaluation of epilepsy. Brain 124(9):1683–1700
Saastamoinen A, Pietilä T, Värri A, Lehtokangas M, Saarinen J (1998) Waveform detection with RBF network-application to automated EEG analysis. Neurocomputing 20(1–3):1–13
San-Segundo R, Gil-Martín M, D’Haro-Enríquez LF, Pardo JM (2019) Classification of epileptic EEG recordings using signal transforms and convolutional neural networks. Comput Biol Med 109:148–158
Sciaraffa N, Klados MA, Borghini G, Di Flumeri G, Babiloni F, Aricò P (2020) Double–step machine learning based procedure for HFOs detection and classification. Brain Sci 10(4)
Sharma R, Pachori RB, Gautam S (2014) Empirical mode decomposition based classification of focal and non-focal seizure EEG signals. In: International conference on medical biometrics (ICMB), pp 135–140
Sharma R, Pachori RB, Acharya UR (2015) Application of entropy measures on intrinsic mode functions for the automated identification of focal electroencephalogram signals. Entropy 17(2):669–691
Sharma R, Pachori RB, Acharya UR (2015) An integrated index for the identification of focal electroencephalogram signals using discrete wavelet transform and entropy measures. Entropy 17(8):5218–5240
Sharma R, Kumar M, Pachori RB, Acharya UR (2017) Decision support system for focal EEG signals using tunable-Q wavelet transform. J Comput Sci 20:52–60
Sharma R, Sircar P, Pachori RB (2020) Automated focal EEG signal detection based on third order cumulant function. Biomed Signal Process Control 58:101856
Shoeb AH (2009) Application of machine learning to epileptic seizure onset detection and treatment. Massachusetts Institute of Technology
Shoeb AH, Guttag JV (2010) Application of machine learning to epileptic seizure detection. In: Proceedings of the 27th International conference on machine learning (ICML), 8, pp 975–982
Siddharth T, Tripathy RK, Pachori RB (2019) Discrimination of focal and non-focal seizures from EEG signals using sliding mode singular spectrum analysis. IEEE Sens J 19(24):12286–12296
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. 1409.1556
Song Y, Zhang J (2013) Automatic recognition of epileptic EEG patterns via extreme learning machine and multiresolution feature extraction. Expert Syst Appl 40(14):5477–5489
Sriraam N, Raghu S (2017) Classification of focal and non focal epileptic seizures using multi-features and svm classifier. J Med Syst 41(10):160
Staba RJ, Wilson CL, Bragin A, Fried I, Engel J (2002) Quantitative analysis of high-frequency oscillations (80–500 hz) recorded in human epileptic hippocampus and entorhinal cortex. J Neurophysiol 88(4):1743–1752
Stafstrom C, Carmant L (2015) Seizures and epilepsy: an overview for neuroscientists. Cold Spring Harbor Perspect Med 5(6)
Staley KJ, Dudek FE (2006) Interictal spikes and epileptogenesis. Epilepsy. Currents 6(6):199–202
Stevens JR, Lonsbury BL, Goel SL (1972) Seizure occurrence and interspike interval: telemetered electroencephalogram studies. Arch Neurol 26(5):409–419
Subasi A, Jukic S, Kevric J (2019) Comparison of EMD, DWT and WPD for the localization of epileptogenic foci using random forest classifier. Measurement 146:846–855
Subathra M, Mohammed MA, Maashi MS et al (2020) Detection of focal and non-focal electroencephalogram signals using fast Walsh-Hadamard transform and artificial neural network. Sensors 20(17):4952
Sui L, Zhao X, Zhao Q, Tanaka T, Cao J (2019) Localization of epileptic foci by using convolutional neural network based on iEEG. In: IFIP International conference on artificial intelligence applications and innovations, Springer, pp 331–339
Talairach J, Bancaud J (1966) Lesion, irritative zone and epileptogenic focus. Confinia Neurol 27(1):91–94
Urrestarazu E, Chander R, Dubeau F, Gotman J (2007) Interictal high-frequency oscillations (100–500 Hz) in the intracerebral EEG of epileptic patients. Brain 130(9):2354–2366
Varatharajah Y, Berry B, Cimbalnik J, Kremen V, Van Gompel J, Stead M, Brinkmann B, Iyer R, Worrell G (2018) Integrating artificial intelligence with real-time intracranial EEG monitoring to automate interictal identification of seizure onset zones in focal epilepsy. J Neural Eng 15(4):046035
Wang J, Hirschmann J, Elben S, Hartmann CJ, Vesper J, Wojtecki L, Schnitzler A (2014) High–frequency oscillations in parkinson’s disease: spatial distribution and clinical relevance. Movement Disorders 29(10)
Warren CP, Hu S, Stead M, Brinkmann BH, Bower MR, Worrell GA (2010) Synchrony in normal and focal epileptic brain: the seizure onset zone is functionally disconnected. J Neurophysiol 104(6):3530–3539
Weiss S, Orosz I, Salamon N, Moy S, Wei L, Klooster MV, Knight R, Harper RM, Bragin A, Fried I Jr, Staba RJE (2016) Ripples on spikes show increased phase-amplitude coupling in mesial temporal lobe epilepsy seizure-onset zones. Epilepsia 57(11):1916–1930
Wilson SB, Emerson R (2002) Spike detection: a review and comparison of algorithms. Clin Neurophysiol 113(12):1873–1881
Winterhalder M, Maiwald T, Voss H, Aschenbrenner-Scheibe R, Timmer J, Schulze-Bonhage A (2003) The seizure prediction characteristic: a general framework to assess and compare seizure prediction methods. Epilepsy Behav 4(3):318–325
Witte H, Eiselt M, Patakova I, Petranek S, Griessbach G, Krajca V, Rother M (1991) Use of discrete Hilbert transformation for automatic spike map**: a methodological investigation. Med Biol Eng Comput 29(3):242–248
Worrell GA, Jerbi K, Kobayashi K, Lina JM, Zelmann R, Le Van Quyen M (2012) Recording and analysis techniques for high-frequency oscillations. Prog Neurobiol 98(3):265–278
Xu G, Wang J, Zhang Q, Zhu J (2006) An automatic EEG spike detection algorithm using morphological filter. In: 2006 IEEE International conference on automation science and engineering, pp 170–175
Xu G, Wang J, Zhang Q, Zhang S, Zhu J (2007) A spike detection method in EEG based on improved morphological filter. Comput Biol Med 37(11):1647–1652
Zelmann R, Mari F, Jacobs J, Zijlmans M, Dubeau F, Gotman J (2012) A comparison between detectors of high frequency oscillations. Clin Neurophysiol 123(1):106–116
Zhao X, Zhao Q, Tanaka T, Cao J, Kong W, Sugano H, Yoshida N (2018) Detection of epileptic foci based on interictal iEEG by using convolutional neural network. In: IEEE 23rd International conference on digital signal processing (DSP), pp 1–5
Zhao X, Solé-Casals J, Zhao Q, Cao J, Tanaka T (2021) Multi-feature fusion for epileptic focus localization based on tensor representation. In: 2021 Asia-Pacific signal and information processing association annual summit and conference (APSIPA ASC), IEEE, pp 1323–1327
Zhu G, Li Y, Wen PP, Wang S, ** M (2013) Epileptogenic focus detection in intracranial EEG based on delay permutation entropy. In: AIP conference proceedings, pp 31–36
Zijlmans M, Jacobs J, Kahn YU, Zelmann R, Dubeau F, Gotman J (2011) Ictal and interictal high frequency oscillations in patients with focal epilepsy. Clin Neurophysiol 122(4):664–671
Zuo R, Wei J, Li X et al (2019) Automated detection of high-frequency oscillations in epilepsy based on a convolutional neural network. Front Comput Neurosci 13:6
Acknowledgements
This work was supported by JST CREST Grant Number JPMJCR1784, Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Islam, M.R., Zhao, X., Miao, Y. et al. Epileptic seizure focus detection from interictal electroencephalogram: a survey. Cogn Neurodyn 17, 1–23 (2023). https://doi.org/10.1007/s11571-022-09816-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11571-022-09816-z