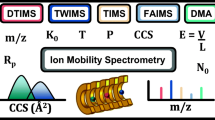
Abstract
Background
Ion mobility (IM) separation capabilities are now widely available to researchers through several commercial vendors and are now being adopted into many metabolomics workflows. The added peak capacity that ion mobility offers with minimal compromise to other analytical figures-of-merit has provided real benefits to sensitivity and structural selectivity and have allowed more specific metabolite annotations to be assigned in untargeted workflows. One of the greatest promises of contemporary IM-enabled instrumentation is the capability of operating multiple analytical dimensions inline with minimal sample volumes, which has the potential to address many grand challenges currently faced in the omics fields. However, comprehensive operation of multidimensional mass spectrometry comes with its own inherent challenges that, beyond operational complexity, may not be immediately obvious to practitioners of these techniques.
Aim of review
In this review, we outline the strengths and considerations for incorporating IM analysis in metabolomics workflows and provide a critical but forward-looking perspective on the contemporary challenges and prospects associated with interpreting IM data into chemical knowledge.
Key scientific concepts of review
We outline a strategy for unifying IM-derived collision cross section (CCS) measurements obtained from different IM techniques and discuss the emerging field of high resolution ion mobility (HRIM) that is poised to address many of the contemporary challenges associated with ion mobility metabolomics. Whereas the LC step limits the throughput of comprehensive LC-IM-MS, the higher peak capacity of HRIM can allow fast LC gradients or rapid sample cleanup via solid-phase extraction (SPE) to be utilized, significantly improving the sample throughput.




Similar content being viewed by others
Abbreviations
- AIF:
-
All Ions Fragmentation
- CCS:
-
Collision Cross Section
- cIM:
-
Cyclic Ion Mobility
- DDA:
-
Data-Dependent Acquisition
- DIA:
-
Data-Independent Acquisition
- DTIMS:
-
Drift Tube Ion Mobility Spectrometry
- FTMS:
-
Fourier Transform Mass Spectrometry
- FWHM:
-
Full Width at Half Maximum
- GC:
-
Gas Chromatography
- HDMS:
-
High Definition Mass Spectrometry
- HRdm:
-
High Resolution Demultiplexing
- HRIM:
-
High Resolution Ion Mobility
- IM:
-
Ion Mobility
- LC:
-
Liquid Chromatography
- m/z:
-
Mass-to-Charge Ratio
- MRT:
-
Multi-Reflecting Time-of-Flight Mass Spectrometry
- MS1:
-
Single Stage Mass Spectrometry
- MS2:
-
Two-Stage Tandem Mass Spectrometry
- MSn :
-
Multiple-Stage (n > 2) Tandem Mass Spectrometry
- PASEF:
-
Parallel Accumulation–Serial Fragmentation
- RT:
-
Retention Time
- SLIM:
-
Structures for Lossless Ion Manipulations
- SPE:
-
Solid-Phase Extraction
- TIMS:
-
Trapped Ion Mobility Spectrometry
- TOFMS:
-
Time-of-Flight Mass Spectrometry
- TWIMS:
-
Traveling Wave Ion Mobility Spectrometry
- TW-SLIM:
-
Traveling Wave Structures for Lossless Ion Manipulations
References
Ali, A., Davidson, S., Fraenkel, E., Gilmore, I., Hankemeier, T., Kirwan, J. A., Lane, A. N., Lanekoff, I., Larion, M., McCall, L. I., Murphy, M., Sweedler, J. V., & Zhu, C. (2022). Single cell metabolism: current and future trends. Metabolomics, 18, 77. https://doi.org/10.1007/s11306-022-01934-3.
Athersuch, T. (2016). Metabolome analyses in exposome studies: profiling methods for a vast chemical space. Archives of biochemistry and biophysics, 589, 177–186. https://doi.org/10.1016/j.abb.2015.10.007.
Breen, J., Hashemihedeshi, M., Amiri, R., Dorman, F. L., & Jobst, K. J. (2022). Unwrap** Wrap-around in gas (or Liquid) Chromatographic Cyclic Ion Mobility–Mass Spectrometry. Analytical Chemistry, 94, 11113–11117. https://doi.org/10.1021/acs.analchem.2c02351.
Causon, T. J., & Hann, S. (2020). Uncertainty estimations for collision cross section determination via uniform field drift tube-ion mobility-mass spectrometry. Journal of the American Society for Mass Spectrometry, 31, 2102–2110. https://doi.org/10.1021/jasms.0c00233.
Causon, T. J., Si-Hung, L., Newton, K., Kurulugama, R. T., Fjeldsted, J., & Hann, S. (2019). Fundamental study of ion trap** and multiplexing using drift tube-ion mobility time-of-flight mass spectrometry for non-targeted metabolomics. Analytical and bioanalytical chemistry, 1–10. https://doi.org/10.1007/s00216-019-02021-8.
Chaleckis, R., Meister, I., Zhang, P., & Wheelock, C. E. (2019). Challenges, progress and promises of metabolite annotation for LC–MS-based metabolomics. Current opinion in biotechnology, 55, 44–50. https://doi.org/10.1016/j.copbio.2018.07.010.
Collins, S. L., Koo, I., Peters, J. M., Smith, P. B., & Patterson, A. D. (2021). Current Challenges and recent developments in Mass Spectrometry–Based Metabolomics. Annual Review of Analytical Chemistry, 14, 467–487. https://doi.org/10.1146/annurev-anchem-091620-015205.
Davis, D. E. Jr., Leaptrot, K. L., Koomen, D. C., May, J. C., Cavalcanti, G. A., Padilha, M. C., Pereira, H. M., & McLean, J. A. (2021). Multidimensional separations of Intact Phase II Steroid Metabolites utilizing LC–Ion Mobility–HRMS. Analytical chemistry, 93, 10990–10998. https://doi.org/10.1021/acs.analchem.1c02163.
de Dias, S., Verbaere, A. L., Meudec, A., Deshaies, E., Saucier, S., Cheynier, C., V. and, & Sommerer, N. (2022). Improved analysis of Isomeric Polyphenol Dimers using the 4th dimension of trapped Ion mobility Spectrometry—Mass Spectrometry. Molecules, 27, 4176. https://doi.org/10.3390/molecules27134176.
Delafield, D. G., Lu, G., Kaminsky, C. J., & Li, L. (2022). High-end Ion Mobility Mass Spectrometry: A Current Review of Analytical Capacity in Omics Applications and Structural Investigations. TrAC Trends in Analytical Chemistry, 116761. https://doi.org/10.1016/j.trac.2022.116761.
Deng, L., Ibrahim, Y. M., Baker, E. S., Aly, N. A., Hamid, A. M., Zhang, X., Zheng, X., Garimella, S. V., Webb, I. K., & Prost, S. A. (2016). Ion mobility separations of isomers based upon long path length structures for lossless ion manipulations combined with mass spectrometry. ChemistrySelect, 1, 2396–2399. https://doi.org/10.1002/slct.201600460.
Dodds, J. N., May, J. C., & McLean, J. A. (2016). Investigation of the complete suite of the leucine and isoleucine isomers: toward prediction of Ion mobility separation capabilities. Analytical Chemistry, 89, 952–959. https://doi.org/10.1021/acs.analchem.6b04171.
Dodds, J. N., May, J. C., & McLean, J. A. (2017). Correlating resolving Power, Resolution, and Collision Cross Section: Unifying Cross-Platform Assessment of separation efficiency in Ion mobility spectrometry. Analytical Chemistry, 89, 12176–12184. https://doi.org/10.1021/acs.analchem.7b02827.
Drakopoulou, S. K., Damalas, D. E., Baessmann, C., & Thomaidis, N. S. (2021). Trapped Ion Mobility Incorporated in LC–HRMS Workflows as an Integral Analytical platform of high sensitivity: targeted and untargeted 4D-Metabolomics in Extra Virgin Olive Oil. Journal of Agricultural and Food Chemistry, 69, 15728–15737. https://doi.org/10.1021/acs.jafc.1c04789.
Feuerstein, M. L., Hernández-Mesa, M., Valadbeigi, Y., Le Bizec, B., Hann, S., Dervilly, G., & Causon, T. (2022). Critical evaluation of the role of external calibration strategies for IM-MS. Analytical and Bioanalytical Chemistry, 414, 7483–7493. https://doi.org/10.1007/s00216-022-04263-5.
Fiehn, O., Barupal, D. K., & Kind, T. (2011). Extending biochemical databases by metabolomic surveys. Journal of Biological Chemistry, 286, 23637–23643. https://doi.org/10.1074/jbc.R110.173617.
Gabelica, V., & Marklund, E. (2018). Fundamentals of ion mobility spectrometry. Current Opinion in Chemical Biology, 42, 51–59. https://doi.org/10.1016/j.cbpa.2017.10.022.
Gabelica, V., Shvartsburg, A. A., Afonso, C., Barran, P., Benesch, J. L., Bleiholder, C., Bowers, M. T., Bilbao, A., Bush, M. F., & Campbell, J. L. et al., (2019). Recommendations for reporting ion mobility Mass Spectrometry measurements. Mass spectrometry reviews, 38, 291–320. https://doi.org/10.1002/mas.21585.
German, J. B., Hammock, B. D., & Watkins, S. M. (2005). Metabolomics: building on a century of biochemistry to guide human health. Metabolomics, 1, 3–9. https://doi.org/10.1007/s11306-005-1102-8.
Giddings, J. C. (1984). Two-dimensional separations: concept and promise. Analytical Chemistry, 56, 1258A–1270. https://doi.org/10.1021/ac00276a003. A.
Giddings, J. C. (1987). Concepts and comparisons in multidimensional separation. Journal of High Resolution Chromatography, 10, 319–323. https://doi.org/10.1002/jhrc.1240100517.
Giles, K., Ujma, J., Wildgoose, J., Pringle, S., Richardson, K., Langridge, D., & Green, M. (2019). A cyclic Ion Mobility-Mass Spectrometry System. Analytical Chemistry, 91, 8564–8573. https://doi.org/10.1021/acs.analchem.9b01838.
Grabarics, M., Lettow, M., Kirk, A. T., von Helden, G., Causon, T. J., & Pagel, K. (2020). Plate-height model of ion mobility-mass spectrometry. The Analyst, 145, 6313–6333. https://doi.org/10.1039/D0AN00433B.
Grabarics, M., Lettow, M., Kirk, A. T., von Helden, G., Causon, T. J., & Pagel, K. (2021). Plate-height model of ion mobility-mass spectrometry: part 2—Peak-to-peak resolution and peak capacity. Journal of Separation Science, 44, 2798–2813. https://doi.org/10.1002/jssc.202100201.
Hernández-Mesa, M., D’atri, V., Barknowitz, G., Fanuel, M., Pezzatti, J., Dreolin, N., Ropartz, D., Monteau, F., Vigneau, E., & Rudaz, S. (2020). Interlaboratory and interplatform study of steroids collision cross section by traveling wave ion mobility spectrometry. Analytical chemistry, 92, 5013–5022. https://doi.org/10.1021/acs.analchem.9b05247.
Hinnenkamp, V., Klein, J., Meckelmann, S. W., Balsaa, P., Schmidt, T. C., & Schmitz, O. J. (2018). Comparison of CCS values determined by traveling wave ion mobility mass spectrometry and drift tube ion mobility mass spectrometry. Analytical chemistry, 90, 12042–12050. https://doi.org/10.1021/acs.analchem.8b02711.
HMDB The Human Metabolome Database, www.hmdb.ca (accessed October 4, 2022).
Ibrahim, Y. M., Hamid, A. M., Deng, L., Garimella, S. V. B., Webb, I. K., Baker, E. S., & Smith, R. D. (2017). New frontiers for mass spectrometry based upon structures for lossless ion manipulations. The Analyst, 142, 1010–1021. https://doi.org/10.1039/C7AN00031F.
Ieritano, C., Le Blanc, Y., Schneider, J., Bissonnette, B. B., Haack, J. R., A. and, & Hopkins, W. S. (2022). Protonation-Induced Chirality drives separation by Differential Ion mobility spectrometry. Angewandte Chemie International Edition, 61, e202116794. https://doi.org/10.1002/anie.202116794.
Kaufmann, A., Butcher, P., Maden, K., Walker, S., & Widmer, M. (2020). Does the ion mobility resolving power as provided by commercially available ion mobility quadrupole time-of-flight mass spectrometry instruments permit the unambiguous identification of small molecules in complex matrices? Analytica Chimica Acta, 1107, 113–126. https://doi.org/10.1016/j.aca.2020.02.032.
Kaufmann, A., Butcher, P., Maden, K., Widmer, M., Giles, K., & Uría, D. (2009). Are liquid chromatography/electrospray tandem quadrupole fragmentation ratios unequivocal confirmation criteria? Rapid Communications in Mass Spectrometry, 23, 985–998. https://doi.org/10.1002/rcm.3959.
Krug, S., Kastenmüller, G., Stückler, F., Rist, M. J., Skurk, T., Sailer, M., Raffler, J., Römisch-Margl, W., Adamski, J., & Prehn, C. (2012). The dynamic range of the human metabolome revealed by challenges. The FASEB Journal, 26, 2607–2619. https://doi.org/10.1096/fj.11-198093.
Lalli, P. M., Iglesias, B. A., Toma, H. E., de Sa, G. F., Daroda, R. J., Silva Filho, J. C., Szulejko, J. E., Araki, K., & Eberlin, M. N. (2012). Protomers: formation, separation and characterization via travelling wave ion mobility mass spectrometry. Journal of Mass Spectrometry, 47, 712–719. https://doi.org/10.1002/jms.2999.
Luo, M. D., Zhou, Z. W., & Zhu, Z. J. (2020). The application of ion mobility-mass spectrometry in untargeted metabolomics: from separation to identification. Journal of Analysis and Testing, 4, 163–174. https://doi.org/10.1007/s41664-020-00133-0.
Mahieu, N. G., & Patti, G. J. (2017). Systems-Level annotation of a Metabolomics Data Set reduces 25 000 features to fewer than 1000 unique metabolites. Analytical Chemistry, 89, 10397–10406. https://doi.org/10.1021/acs.analchem.7b02380.
May, J. C., Knochenmuss, R., Fjeldsted, J. C., & McLean, J. A. (2020). Resolution of Isomeric Mixtures in Ion mobility using a combined demultiplexing and peak deconvolution technique. Analytical Chemistry, 92, 9482–9492. https://doi.org/10.1021/acs.analchem.9b05718.
May, J. C., Leaptrot, K. L., Rose, B. S., Moser, K. L. W., Deng, L., Maxon, L., DeBord, D., & McLean, J. A. (2021). Resolving power and Collision Cross Section Measurement Accuracy of a Prototype High-Resolution Ion mobility platform incorporating structures for Lossless Ion Manipulation. Journal of the American Society for Mass Spectrometry, 32, 1126–1137. https://doi.org/10.1021/jasms.1c00056.
May, J. C., & McLean, J. A. (2015). Ion Mobility-Mass Spectrometry: Time-Dispersive Instrumentation. Analytical Chemistry, 87, 1422–1436. https://doi.org/10.1021/ac504720m.
May, J. C., & McLean, J. A. (2016). Advanced Multidimensional Separations in Mass Spectrometry: Navigating the Big Data Deluge. Annual Review of Analytical Chemistry9. https://doi.org/10.1146/annurev-anchem-071015-041734
May, J. C., Morris, C. B., & McLean, J. A. (2017). Ion mobility Collision Cross Section Compendium. Analytical Chemistry, 89, 1032–1044. https://doi.org/10.1021/acs.analchem.6b04905.
Michelmann, K., Silveira, J. A., Ridgeway, M. E., & Park, M. A. (2015). Fundamentals of Trapped Ion mobility spectrometry. Journal of The American Society for Mass Spectrometry, 26, 14–24. https://doi.org/10.1007/s13361-014-0999-4.
Moser, K. L. W., Van Aken, G., DeBord, D., Hatcher, N. G., Maxon, L., Sherman, M., Yao, L., & Ekroos, K. (2021). High-defined quantitative snapshots of the ganglioside lipidome using high resolution ion mobility SLIM assisted shotgun lipidomics. Analytica Chimica Acta, 1146, 77–87. https://doi.org/10.1016/j.aca.2020.12.022.
Nichols, C. M., Dodds, J. N., Rose, B. S., Picache, J. A., Morris, C. B., Codreanu, S. G., May, J. C., Sherrod, S. D., & McLean, J. A. (2018). Untargeted Molecular Discovery in primary metabolism: Collision Cross Section as a Molecular Descriptor in Ion Mobility-Mass Spectrometry. Analytical chemistry, 90, 14484–14492. https://doi.org/10.1021/acs.analchem.8b04322.
Ogata, K., & Ishihama, Y. (2020). Extending the separation space with trapped ion mobility spectrometry improves the accuracy of isobaric tag-based quantitation in proteomic LC/MS/MS. Analytical Chemistry, 92, 8037–8040. https://doi.org/10.1021/acs.analchem.0c01695.
Paglia, G., & Astarita, G. (2017). Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry. Nature protocols, 12, 797–813. https://doi.org/10.1038/nprot.2017.013.
Paglia, G., Smith, A. J., & Astarita, G. (2022). Ion mobility mass spectrometry in the omics era: Challenges and opportunities for metabolomics and lipidomics. Mass Spectrometry Reviews, 41, 722–765. https://doi.org/10.1002/mas.21686.
Picache, J. A., May, J. C., & McLean, J. A. (2020). Crowd-sourced chemistry: considerations for building a standardized database to improve omic analyses. ACS omega, 5, 980–985. https://doi.org/10.1021/acsomega.9b03708.
Picache, J. A., Rose, B. S., Balinski, A., Leaptrot, K. L., Sherrod, S. D., May, J. C., & McLean, J. A. (2019). Collision Cross Section Compendium to annotate and predict multi-omic compound identities. Chemical Science, 983–993. https://doi.org/10.1039/C8SC04396E.
Rappaport, S. M., Barupal, D. K., Wishart, D., Vineis, P., & Scalbert, A. (2014). The blood exposome and its role in discovering causes of disease. Environmental health perspectives, 122, 769–774. https://doi.org/10.1289/ehp.1308015.
Regueiro, J., Negreira, N., & Berntssen, M. H. (2016). Ion-mobility-derived collision cross section as an additional identification point for multiresidue screening of pesticides in fish feed. Analytical chemistry, 88, 11169–11177. https://doi.org/10.1021/acs.analchem.6b03381.
Ridgeway, M. E., Lubeck, M., Jordens, J., Mann, M., & Park, M. A. (2018). Trapped ion mobility spectrometry: a short review. International Journal of Mass Spectrometry, 425, 22–35. https://doi.org/10.1016/j.ijms.2018.01.006.
Rister, A. L., & Dodds, E. D. (2020). Steroid analysis by ion mobility spectrometry. Steroids, 153, 108531. https://doi.org/10.1016/j.steroids.2019.108531.
Rose, B. S., Leaptrot, K. L., Harris, R. A., Sherrod, S. D., May, J. C., & McLean, J. A. (2021). High confidence shotgun lipidomics using structurally selective ion mobility-mass spectrometry, Mass Spectrometry-Based Lipidomics, Springer. pp. 11–37. https://doi.org/10.1007/978-1-0716-1410-5_2
Rose, B. S., May, J. C., Picache, J. A., Codreanu, S. G., Sherrod, S. D., & McLean, J. A. (2022). Improving confidence in lipidomic annotations by incorporating empirical ion mobility regression analysis and chemical class prediction. Bioinformatics, 38, 2872–2879. https://doi.org/10.1093/bioinformatics/btac197.
Scalbert, A., Brennan, L., Manach, C., Andres-Lacueva, C., Dragsted, L. O., Draper, J., Rappaport, S. M., van der Hooft, J. J., & Wishart, D. S. (2014). The food metabolome: a window over dietary exposure. The American journal of clinical nutrition, 99, 1286–1308. https://doi.org/10.3945/ajcn.113.076133.
Schmitt-Kopplin, P., Hemmler, D., Moritz, F., Gougeon, R. D., Lucio, M., Meringer, M., Müller, C., Harir, M., & Hertkorn, N. (2019). Systems chemical analytics: introduction to the challenges of chemical complexity analysis. Faraday Discussions, 218, 9–28. https://doi.org/10.1039/C9FD00078J.
Shliaha, P. V., Bond, N. J., Gatto, L., & Lilley, K. S. (2013). Effects of traveling Wave Ion mobility separation on Data Independent Acquisition in Proteomics Studies. Journal of Proteome Research, 12, 2323–2339. https://doi.org/10.1021/pr300775k.
Silveira, J. A., Danielson, W., Ridgeway, M. E., & Park, M. A. (2016). Altering the mobility-time continuum: nonlinear scan functions for targeted high resolution trapped ion mobility-mass spectrometry. International Journal for Ion Mobility Spectrometry, 19, 87–94. https://doi.org/10.1007/s12127-016-0196-1.
Stow, S. M., Causon, T. J., Zheng, X., Kurulugama, R. T., Mairinger, T., May, J. C., Rennie, E. E., Baker, E. S., Smith, R. D., McLean, J. A., Hann, S., & Fjeldsted, J. C. (2017). An interlaboratory evaluation of Drift Tube Ion Mobility-Mass Spectrometry Collision Cross Section measurements. Analytical Chemistry, 89, 9048–9055. https://doi.org/10.1021/acs.analchem.7b01729.
Tejada-Casado, C., Hernández-Mesa, M., Monteau, F., Lara, F. J., del Olmo-Iruela, M., García-Campaña, A. M., Le Bizec, B., & Dervilly-Pinel, G. (2018). Collision cross section (CCS) as a complementary parameter to characterize human and veterinary drugs. Analytica chimica acta, 1043, 52–63. https://doi.org/10.1016/j.aca.2018.09.065.
Uppal, K., Walker, D. I., Liu, K., Li, S., Go, Y. M., & Jones, D. P. (2016). Computational metabolomics: a framework for the million metabolome. Chemical research in toxicology, 29, 1956–1975. https://doi.org/10.1021/acs.chemrestox.6b00179.
Vasilopoulou, C. G., Sulek, K., Brunner, A. D., Meitei, N. S., Schweiger-Hufnagel, U., Meyer, S. W., Barsch, A., Mann, M., & Meier, F. (2020). Trapped ion mobility spectrometry and PASEF enable in-depth lipidomics from minimal sample amounts. Nature communications, 11, 1–11. https://doi.org/10.1038/s41467-019-14044-x.
Wasito, H., Causon, T., & Hann, S. (2022). Alternating in-source fragmentation with single-stage high-resolution mass spectrometry with high annotation confidence in non-targeted metabolomics. Talanta, 236, 122828. https://doi.org/10.1016/j.talanta.2021.122828.
Wu, Q., Wang, J. Y., Han, D. Q., & Yao, Z. P. (2020). Recent advances in differentiation of isomers by ion mobility mass spectrometry. TrAC Trends in Analytical Chemistry, 124, 115801. https://doi.org/10.1016/j.trac.2019.115801.
**a, J., **ao, W., Lin, X., Zhou, Y., Qiu, P., Si, H., Wu, X., Niu, S., Luo, Z., & Yang, X. (2022). Ion mobility-derived collision cross-sections add Extra Capability in distinguishing Isomers and Compounds with similar Retention Times: the case of Aphidicolanes. Marine Drugs, 20, 541. https://doi.org/10.3390/md20090541.
Xu, Z., Li, J., Chen, A., Ma, X., & Yang, S. (2018). A new retrospective, multi-evidence veterinary drug screening method using drift tube ion mobility mass spectrometry. Rapid Communications in Mass Spectrometry, 32, 1141–1148. https://doi.org/10.1002/rcm.8154.
Zenobi, R. (2013). Single-cell metabolomics: analytical and biological perspectives. Science, 342, 1243259. https://doi.org/10.1126/science.1243259.
Zhang, X., Romm, M., Zheng, X., Zink, E. M., Kim, Y. M., Burnum-Johnson, K. E., Orton, D. J., Apffel, A., Ibrahim, Y. M., Monroe, M. E., Moore, R. J., Smith, J. N., Ma, J., Renslow, R. S., Thomas, D. G., Blackwell, A. E., Swinford, G., Sausen, J., Kurulugama, R. T., Eno, N., Darland, E., Stafford, G., Fjeldsted, J., Metz, T. O., Teeguarden, J. G., Smith, R. D., & Baker, E. S. (2016). SPE-IMS-MS: an automated platform for sub-sixty second surveillance of endogenous metabolites and xenobiotics in biofluids. Clinical Mass Spectrometry, 2, 1–10. https://doi.org/10.1016/j.clinms.2016.11.002.
Zheng, X., Aly, N. A., Zhou, Y., Dupuis, K. T., Bilbao, A., Paurus, V. L., Orton, D. J., Wilson, R., Payne, S. H., & Smith, R. D. (2017). A structural examination and collision cross section database for over 500 metabolites and xenobiotics using drift tube ion mobility spectrometry. Chemical Science, 8, 7724–7736. https://doi.org/10.1039/C7SC03464D.
Valentina, Calabrese Isabelle, Schmitz-Afonso Candice, Prevost Carlos, Afonso Abdelhakim, Elomri (2022) Molecular networking and collision cross section prediction for structural isomer and unknown compound identification in plant metabolomics: a case study applied to Zhanthoxylum heitzii extracts. Analytical and Bioanalytical Chemistry 414(14) 4103-4118 10.1007/s00216-022-04059-7
Zhiwei, Zhou Mingdu, Luo **, Chen Yandong, Yin **n, **ong Ruohong, Wang Zheng-Jiang, Zhu (2020) Ion mobility collision cross-section atlas for known and unknown metabolite annotation in untargeted metabolomics. Nature Communications 11(1) 4334 10.1038/s41467-020-18171-8
Acknowledgements
This work was supported in part using the resources of the Center for Innovative Technology (CIT) at Vanderbilt University. Financial support for aspects of this work was provided by the U.S. Environmental Protection Agency (EPA) under grant No. R839504. This work has not been formally reviewed by the EPA and EPA does not endorse any products or commercial services mentioned in this document. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the EPA or the U.S. Government.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to this work.
Corresponding author
Ethics declarations
Statements and declarations
The authors acknowledge collaborative agreements with Waters Corporation, Agilent Technologies, and MOBILion Systems, which are manufacturers of ion mobility instrumentation. JAM is a member of the Scientific Advisory Board for MOBILion Systems. The authors certify that their contributions are scientifically objective and are not influenced by these collaborative agreements or SAB participation.
Competing interests
The authors acknowledge collaborative agreements with Waters Corporation, Agilent Technologies, and MOBILion Systems, which are manufacturers of ion mobility instrumentation. JAM is a member of the Scientific Advisory Board for MOBILion Systems. The authors certify that their contributions are scientifically objective and are not influenced by these collaborative agreements or SAB participation.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
May, J.C., McLean, J.A. Integrating ion mobility into comprehensive multidimensional metabolomics workflows: critical considerations. Metabolomics 18, 104 (2022). https://doi.org/10.1007/s11306-022-01961-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-022-01961-0