Abstract
The gut microbiota is a central regulator of host metabolism. The composition and function of the gut microbiota is dynamic and affected by diet properties such as the amount and composition of lipids. Hence, dietary lipids may influence host physiology through interaction with the gut microbiota. Lipids affect the gut microbiota both as substrates for bacterial metabolic processes, and by inhibiting bacterial growth by toxic influence. The gut microbiota has been shown to affect lipid metabolism and lipid levels in blood and tissues, both in mice and humans. Furthermore, diseases linked to dyslipidemia, such as non-alcoholic liver disease and atherosclerosis, are associated with changes in gut microbiota profile. The influence of the gut microbiota on host lipid metabolism may be mediated through metabolites produced by the gut microbiota such as short-chain fatty acids, secondary bile acids and trimethylamine and by pro-inflammatory bacterially derived factors such as lipopolysaccharide. Here we will review the association between gut microbiota, dietary lipids and lipid metabolism
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The gut microbiota regulates many metabolic processes in the host including energy homeostasis, glucose metabolism and lipid metabolism [1]. Microbial imbalance, sometimes termed dysbiosis, is associated with metabolic perturbations, and several studies have demonstrated a causal relationship between microbial function and metabolic perturbations. Therapies targeted against the gut microbiota have been shown to improve the metabolic function in humans [2, 3] and transplantation of the fecal microbiota from patients with obesity, steatosis or type 2 diabetes can partly reproduce the donor’s metabolic phenotype in mouse recipients [4,5,6,7].
Lipid metabolism includes the biosynthesis and degradation of lipids such as fatty acids, triglycerides and cholesterol. Specialized lipoproteins facilitate the transport of lipids from the gut to the liver (the site of most lipid transformations) and between the liver and peripheral tissues. Obesity is linked to dysregulation of lipid metabolism, which may result in abnormal levels of blood lipids, ectopic lipid deposition and associated metabolic diseases such as non-alcoholic liver disease (NAFLD) [8] and atherosclerosis [9]. Lipid metabolism is mainly regulated by nutrients such as sugars and fatty acids. However, several reports have shown that lipid levels are associated with the gut microbiota composition, and mechanistic links between lipid metabolism and microbial metabolites have been described in mouse models.
The gut microbiota has the capacity to perform many processes that cannot be carried out by the host. These processes can give rise to microbially produced or modulated metabolites that function as metabolic substrates and signaling molecules in the host, with major implications for host metabolism and health. Dietary composition is central to the metabolic output of the gut microbiota because: (1) the gut microbiota processes dietary nutrients into metabolites and (2) the diet affects the gut microbiota composition and thereby its metabolic potential and impact on the host. In particular, the importance of dietary fibers for gut microbiota composition and function has been extensively studied. In addition, several studies have reported an important role for dietary lipids.
In this review, we will discuss interactions between lipids, the gut microbiota and the host. We will describe the current knowledge on how dietary lipids affect the gut microbiota, how interactions between dietary lipids and the gut microbiota influence host physiology and health, and how the gut microbiota affects host lipid metabolism.
2 Interaction between dietary lipids and the gut microbiota
The gut microbiota has been shown to differ between mice fed diets that are high or low in fat and between diets that contain equal amounts of fat but from different sources [10,11,12,13] (Fig. 1). A comparison of mice on a variety of diets (low-fat diet and diets containing high levels of saturated fat, n-6 PUFA or n-3 PUFA) showed that diets with saturated fat or n-6 PUFA induced weight gain, but only saturated fat increased insulin resistance, colonic permeability, and mesenteric fat inflammation [12]. The gut microbiota composition of mice fed a low-fat diet and a n-3 PUFA diet differed from the other groups, whereas the gut microbiota of mice fed a saturated fat diet and a n-6 PUFA diet were similar. In another study where mice fed a high-fat diet containing lard, rich in saturated fat, were compared to mice fed a isocaloric high-fat diet containing fish oil, rich in n-3 PUFA, it was found that phylogenetic diversity and abundance of the beneficial bacteria Akkermansia muciniphila, Lactobacillus and Bifidobacterium were lower in mice on a lard diet [10]. The lard diet also reduced insulin sensitivity and increased inflammation in white adipose tissue (WAT) through activation of Toll-like receptor 4 (TLR4) signaling. Transplantation of cecum microbiota into germ-free (GF) mice showed that the diet-induced differences in host phenotype were partly caused by the gut microbiota.
Interaction between dietary lipids and the gut microbiota influences host physiology. Free fatty acids are produced from lipid precursors by the action of lipases. Fatty acids may have antibacterial activity or may be utilized as metabolic substrates by gut bacteria, thereby affecting gut microbiota profile and production of microbial metabolites. This may influence host physiology and health
Not only fat sources with major differences in lipid composition, such as lard and fish oil, but also fat sources that are more similar, may give rise to different gut microbiota composition and function. Devkota et al showed that a diet with milk fat, but not diets with lard or safflower oil, increased expansion of Bilophila wadsworthia in mice [13]. Milk fat promoted taurine-conjugation of bile acids, which increases the availability of sulfur used by B. wadsworthia. The increased levels of B. wadsworthia were associated with a pro-inflammatory immune response and increased incidence of colitis in genetically susceptible mice. In another study that combined lard or palm oil with dietary bile acids, lard enhanced fat mass accumulation, impaired glucose tolerance, and elevated levels of hepatic triglycerides in conventionally raised (CONV-R) but not in GF mice when compared to palm oil [11]. The lard diet also promoted a shift in gut microbiota composition and functions, including changes with potential impact on lipid and amino acid metabolism.
The mechanisms by which dietary fatty acids affect gut microbiota are not well defined. Although most of the fatty acids consumed are absorbed in the small intestine, a minority will pass through the gastrointestinal tract and may therefore directly modulate colonic microbiota composition. Fatty acids have a broad spectrum of antibacterial activity including lysis and solubilization of bacterial cell membranes [14, 15] and inhibition of ATP production [16]. The antibacterial action of fatty acids is affected by carbon chain length, saturation and double bond position [17]. However, the impact of fatty acids on the gut microbiota is not limited to antibiotic action. Although gut anaerobes cannot produce energy by beta-oxidation, bacteria can metabolize fatty acids through other pathways. For example, in a mouse model of alcoholic liver disease, ethanol was found to inhibit biosynthesis of saturated fatty acids by the intestinal microflora. Dietary supplementation with saturated long-chain fatty acids, which were metabolized by and promoted growth of Lactobacillus, reversed alcohol-induced dysbiosis, stabilized the intestinal gut barrier, and reduced liver injury [18].
Intestinal bacteria can also react with fatty acid double bonds to produce metabolites that cannot be synthesized by mammalian hosts. Bacterial processing of linoleic acid, for example, has been shown to produce metabolites that may influence host physiology and health. Conjugated linoleic acid (CLA) can be produced by several gut bacteria including Lactobacillus, Butyrivibrio, and Megasphaera [19, 20]. Different CLA isomers have been demonstrated to have different, and sometimes opposite, effects on the host: c9,t11-CLA improves insulin sensitivity and decreases atherosclerosis by activation of proliferator-activated receptor γ (PPARγ), t10,c12-CLA worsens insulin sensitivity and atherosclerosis by inhibiting expression of PPARγ and LXRα [21,22,23], and t9,t11-CLA reduces atherosclerosis by activation of LXRα [24]. Different bacteria produce different ratios of CLA isomers [25, 26] and promotion of bacteria that produce high levels of beneficial CLAs could therefore potentially be used to promote a healthy metabolic phenotype.
Bacterial production of CLAs is a multistep process involving several metabolic intermediates. These metabolites include several hydroxy fatty acids that affect processes related to host health. 10-hydroxy-cis-12-octadecenoic acid (HYA) enhances intestinal barrier function and suppresses the development of colitis in mice in a free fatty acids 1 (FFR1/GPR40)-dependent manner [27]. Another hydroxylated CLA intermediate - 10-oxo-cis-12-octadecenoic acid (KetoA) - increases adiponectin production and glucose uptake in a PPARγ-dependent manner, and contributes to the prevention of obesity-related metabolic perturbations [28].
3 Influence of the gut microbiota on host lipid metabolism and pathophysiology
Studies in gnotobiotic mice have shown that the gut microbiota affects host lipid metabolism. Importantly, GF mice are protected against diet-induced obesity through a combination of several mechanisms including increased fatty acid oxidation and decreased deposition of triglycerides in adipocytes compared to CONV-R mice [29]. Furthermore, lipidomics analysis of GF and CONV-R mice fed a regular chow diet showed that the gut microbiota affects lipid composition in host tissues and serum and increases clearance of triglycerides from the circulation [30]. In contrast, circulating triglycerides, HDL, and total cholesterol levels are increased by the gut microbiota in mice on a high-fat diet [31]. Comparisons between CONV-R and GF mice have also shown that the gut microbiota induces hepatic production of monounsaturated fatty acids and elongation of PUFA, and that acetate produced by the gut microbiota is used as precursor in hepatic fatty acid synthesis [32].
The gut microbiota affects host lipid metabolism and lipid composition through interaction with the diet. In a recent study by Just et al, CONV-R and GF mice were fed palm oil or lard diet (both rich in saturated lipids) supplemented with bile acids [11]. They found that the gut microbiota increased hepatic triglycerides and cholesteryl esters levels only in mice fed lard, and that colonization status had a major impact on hepatic lipids. In another study where CONV-R and GF mice were fed lard or fish oil diet it was also found that the microbiota downregulates cholesterol biosynthesis and increased hepatic levels of cholesterol specifically in mice fed lard [33]. However, in this study the relative contribution of the gut microbiota to the total variation in the hepatic lipid dataset was small and no serum lipids differed significantly between CONV-R and GF mice. This discrepancy between the two studies could possibly be attributed to the relative similarity between palm oil and lard compared to fish oil and lard.
Studies in mice treated with probiotics provide further evidence for a role of the gut microbiota in regulation of host lipid homeostasis. In mice fed a high-fat high-cholesterol diet, Lactobacillus curvatus alone or together with Lactobacillus plantarum reduced cholesterol in plasma and liver and the two strains had a synergistic effect on hepatic triglycerides [34]. Similarly, in obese rats fed a high-fat diet, Bifidobacterium spp. decreased levels of circulating triglycerides and LDL and increased levels of HDL [35].
Overall, studies in mouse models show that the gut microbiota, in concert with the diet, regulates host lipid metabolism and lipid levels in serum and tissues.
The fecal microbiota has also been linked to lipid metabolism in humans. Taxonomy and functional profiles of the bacteria differ between obese and lean subjects, but results from different studies are inconsistent, in part because of the complex nature of obesity but also because different methods have been used to analyze the microbiota [36]. A number of studies have investigated the association between the gut microbiota and dyslipidemia. When profiling metagenomics data from obese individuals, Cortillard et al found that reduced total microbial gene richness was associated with increased total serum cholesterol and serum triglycerides in obese patients. An energy-restricted diet intervention increased microbial gene richness and reduced serum lipids [37]. Similarly, Le Chantelier et al showed that triglycerides were higher and HDL levels were lower in individuals with low microbial gene counts than in those with high microbial gene counts [38]. Furthermore, by performing cross-validation analysis on taxonomic data of fecal microbiota, biometrics and metabolic measurements from a general population cohort study in the Netherlands, Fu et al could attribute 6% of variance of serum triglycerides and 4% in HDL to the gut microbiota composition [39].
Changes in fecal microbiota composition are also present in individuals with pathophysiological conditions associated with dyslipidemia and ectopic fat deposition such as atherosclerosis and fatty liver. By analyzing the microbial composition of atherosclerotic plaques, fecal samples and the oral cavity in patients with symptomatic carotid artery stenosis [40], Koren et al observed shared operational taxonomic units (OTUs) between all three sites, consistent with the possibility that oral and gastrointestinal microbiota might be involved in inflammatory processes responsible for atherosclerosis. OTUs attributed to Fusobacterium from the oral cavity correlated with total serum cholesterol and LDL, and Streptococcus OTUs correlated with HDL levels. However, there was no clear fecal microbial signature that could distinguish between patients and controls [40]. In contrast, Karlsson et al showed that individuals with symptomatic atherosclerosis had higher abundance of the genus Collinsella and lower abundance of Eubacterium and Roseburia compared with healthy controls. Functionally, the gut microbiome of patients had increased capacity for peptidoglycan synthesis, which might contribute to the chronic inflammation in the atherosclerotic arterial walls [41]. Another study found an association between coronary artery disease (CAD) and gut microbiota composition by demonstrating that the phylum Bacteroidetes was decreased and the order Lactobacillales was increased in CAD patients compared to both subjects with coronary risk factors but without CAD and healthy controls [42].
Several studies have shown that the fecal microbiota composition in subjects with NAFLD differs from that of healthy controls and obese patients without fatty liver disease [43,44,45]. Hoyles et al recently show that patients with steatosis have decreased microbial gene richness and altered genetic potential for several functions including the processing of dietary lipids [46]. Furthermore, changes in fecal microbiota composition have been shown to associate with the severity of NAFLD and its progression to fibrosis and non-alcoholic steatohepatitis (NASH) [47, 48]. An increased abundance of Bacteroidetes in patients with fibrosis or NASH is the most consistent finding in these studies.
4 Mechanisms linking gut microbiota and host lipid metabolism
4.1 Short-chain fatty acids
Short-chain fatty acids (SCFAs) such as acetate, propionate and butyrate are bacterial metabolites derived from fermentation of fibers in the colon (Fig. 2). Both butyrate and propionate have low systemic concentrations whereas acetate levels are higher [49]. SCFAs are important for host metabolism and are used as substrates for energy production, lipogenesis, gluconeogenesis and cholesterol synthesis [50, 51]. Butyrate is an energy source for colonocytes while propionate is mainly metabolized by the liver.
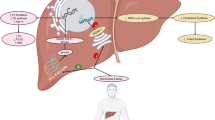
Mechanisms linking the gut microbiota to lipid metabolism and pathophysiological conditions associated with dyslipidemia. Short-chain fatty acids regulate host lipid metabolism by supplying the host with energy, improving peripheral tissue metabolism and stimulating incretin hormone production. The gut microbiota transforms choline and L-carnitine to trimethylamine (TMA). TMA is transformed into trimethylamine N-oxide (TMAO) that may promote increased atherosclerosis through mechanisms related to lipid metabolism and inflammation. Bile acids regulate metabolism by binding to farnesoid X receptor (FXR) and G protein-coupled bile acid receptor 1 (TGR5) in several different tissues. Deconjugation of bile acids reduces absorption and increase excretion of bile acids. Increased gut permeability facilitates translocation of lipopolysaccharide (LPS) over the intestinal epithelium. LPS induce inflammation through TLR4 that may result in metabolic perturbations and contribute to development of metabolic diseases. HDL may neutralize the toxic effect of LPS. PPARγ, peroxisome proliferator-activated receptor gamma; GPR43, G-protein coupled receptors GPR43(FFAR2); GPR41, G-protein coupled receptors GPR43(FFAR3); SCFA, short-chain fatty acid; PYY, peptide YY; TLR4, toll-like receptor 4
In addition to being metabolic substrates, SCFAs act as signaling molecules, notably through the G-protein coupled receptors GPR43/FFAR2 and GPR41/FFAR3. GPR43 protects against diet-induced-obesity in mice [52,53,54,55]. Activation of GPR43 on L-cells increases secretion of glucagon-like peptide-1 (GLP-1) [52, 56] and acetate induces anti-lipolytic activity [57] and improves glucose and lipid metabolism [53] through GPR43 in WAT. GRP41 has also been shown to regulate metabolism through interaction with the gut microbiota. CONV-R Gpr41 knockout mice are leaner and weigh less than their wild-type littermates, while these differences are not found in GF mice. Furthermore, the microbiota increases peptide YY (PYY) production through GPR41 [58]. Butyrate and propionate have also been shown to activate PPARγ [59], and SCFA-induced activation of PPARγ modulates lipid metabolism through increased energy expenditure [60], reduced body weight and decreased liver triglyceride accumulation [61].
Overall, SCFAs have been shown to have a positive impact on metabolic health [62]. Supplementation with acetate reduces weight gain and improves glucose tolerance in obese and diabetic rats [63], butyrate protects against obesity and increases thermogenesis in mice [60] and propionate or butyrate improves glucose homeostasis in mice [64]. Some minor clinical trials have also found beneficial effects of SCFA or fiber supplementation on body weight [56, 65]. Ingestion of a propionate precursor increases postprandial plasma PYY and GLP-1 and reduces energy intake while long-term treatment results in a reduction in weight gain [56]. Plasma concentrations of PYY and GLP-1 are also increased by acetate in humans [66].
4.2 Bile acids
Primary bile acids are synthesized from cholesterol and conjugated to taurine or glycine in the liver. The bile acids are stored in the gallbladder and excreted into the duodenum after food ingestion to aid emulsification of dietary lipids. Most of the bile acids are reabsorbed and recirculated to the liver, but bacterially mediated deconjugation of the glycine or taurine group reduces reabsorption. Deconjugated bile acids can be further metabolized to secondary bile acids through dehydrogenation, dehydroxylation and epimerization by colonic bacteria [67]. Microbial processing results in a more hydrophobic bile acid pool and facilitates excretion in the feces. Fecal excretion of bile acids is a major sink for cholesterol and bile acids lost in the process need to be replaced by de novo synthesis from cholesterol [67].
In addition to their role in lipid digestion, bile acids can act as signaling molecules that regulate host metabolism by binding to the nuclear receptor farnesoid X receptor (FXR) and the Takeda G-protein coupled bile acid receptor TGR5. Microbial processing of bile acids increases the diversity of the bile acid pool and the different bile acids vary in their affinity to the receptors and can act as agonists or antagonists. Both of the primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA) and the secondary bile acids lithocholic acid (LCA) and deoxycholic acid (DCA) are FXR agonists, but with different affinities [68]. In humans, CDCA can be transformed into ursodeoxycholic acid (UDCA), which is a FXR antagonist [69]. Furthermore, the taurine-conjugated murine bile acid TβMCA, but not its deconjugated counterpart βMCA, is a potent FXR antagonist [70].
FXR is involved in the regulation of lipid metabolism, especially triglyceride trafficking, synthesis and utilization [71]. Microbial processing of bile acids may therefore influence lipid metabolism through interaction with FXR. By feeding wild-type and Fxr knockout mice with or without bacteria a high-fat diet, Parséus et al showed that microbiota-induced weight gain, steatosis and inflammation were dependent on FXR signaling [72]. FXR also changed the gut microbiota composition, and transplantation of the gut microbiota into GF mice transferred the lean phenotype of Fxr knockout donor mice, demonstrating that FXR may contribute to increased adiposity by altering the microbiota composition. Comparison of whole body and tissue-specific Fxr knockout mice have revealed that activation of the liver and intestinal FXR result in distinct metabolic outcomes in obesity models [73,74,89, 90] may disrupt the gut barrier function allowing translocation of LPS [91, 92]. This results in moderately increased levels of LPS in the blood which is defined as metabolic endotoxemia [91], a condition linked to metabolic perturbations such as dyslipidemia, insulin resistance, NAFLD and cardiovascular disease [93].
LPS interacts with blood lipids in various ways. First, it increases the concentration of blood triglycerides by multiple mechanisms. In rats, low-dose LPS increases hepatic synthesis of VLDL, whereas high-dose LPS decreases lipoprotein catabolism [94]. Mice lacking the TLR4 co-receptor CD14 are resistant to hyperinsulinemia, insulin resistance and steatosis induced by a high-fat diet or LPS [91]. Second, plasma lipoproteins, in particular HDL, have the ability to neutralize the toxic effects of LPS [95, 96]. The capacity of HDL to bind LPS may protect against inflammation. This is supported by the observations that infusion of HDL prior to a LPS challenge reduced release of proinflammatory cytokines in humans [97] and that LPS induces higher levels of TNFα in hypolipidemic rats compared with controls [98].
LPS has been shown to promote atherosclerosis and cardiovascular disease. LPS-treated hypercholesterolemic rabbits have increased atherosclerosis compared with controls [99] and mice lacking TLR4 have reduced atherosclerosis and plaques with decreased amounts of lipid [100]. In humans, high LPS levels during chronic infections are predictors of increased atherosclerotic risk [101] and metabolic endotoxemia increases the risk for cardiovascular disease and mortality in patients with chronic kidney disease [102]. Human TLR4 mutations have been shown to be associated with a decreased response to LPS [103], reduced risk of carotid artery atherosclerosis [104] and acute coronary events [105].
Mouse studies have shown that hepatic steatosis is induced by a high-fat diet and associated with dysbiosis and increased intestinal permeability [91]. Moreover, chemically induced colitis in rats increases the levels of circulating LPS and worsens steatohepatitis during high-fat diet[106]. Dysbiosis-induced permeability increases the levels of TLR ligands in the portal vein, thereby activating hepatic Kupffer cells and stellate cells to stimulate pro-inflammatory and pro-fibrotic pathways via inflammatory cytokines[107, 108]. In addition, mucosal TLR activation appears to contribute to hepatic steatosis via the TLR adaptor MYD88 expressed in the intestine[109]. Mice with an intestinal epithelial cell-specific deletion of MYD88 fed a high-fat diet have improved glucose homeostasis and decreased hepatic lipid content compared with wild-type mice[110]. TLR signaling in the mucosa can also induce production of inflammasomes, multiprotein oligomers responsible for the activation of inflammatory responses. Inflammasomes activate a variety of pro-inflammatory and pro-fibrotic processes involved in the progression of liver disease[111]. For example, activation of the NLRP3-inflammasome by LPS via TLR4 and TLR9 is involved in the development of fibrosis in NAFLD[112]. Gut permeability has also been linked to NAFLD in humans. NAFLD patients have been shown to have increased gut permeability compared to healthy controls and gut permeability correlated with severity of steatosis but not with steatohepatitis in patients with NAFLD[113]. Interestingly, patients with steatosis have also been shown to have a gut microbiota with increased genetic potential for biosynthesis of endotoxin[46].
4.4 Trimethylamine/Trimethylamine N-oxide
The gut microbiota metabolizes methylamine-containing nutrients such as choline, lecithin and L-carnitine to generate trimethylamine (TMA), which is further processed to trimethylamine N-oxide (TMAO) by flavin monooxygenases (FMO) in the liver. TMAO levels have been correlated with risk of cardiovascular events[114] and prevalence of cardiovascular disease[115, 116]. Plasma TMAO levels in different mouse strains have been positively correlated with lesion size [117] and transfer of microbiota from high- and low-TMAO-producing mice to atherosclerosis-prone Apoe knockout mice show that increased microbial capacity for TMA production increases aortic lesions[118].
FMO3 is the primary enzyme converting TMA into TMAO. Knockdown of Fmo3 results in reduced atherosclerotic lesion areas, altered lipid and cholesterol metabolism, and decreased TMAO plasma levels [119, 120]. FMO3 expression is regulated by bile acids by a mechanism that involves FXR [121]. Gut microbiota processing of bile acids could therefore be an alternative mechanism by which the gut microbiota regulates TMAO production.
The mechanisms by which TMAO contributes to atherosclerosis appears to be complex and not fully understood. Antibiotic treatment reduces production of TMA and has been shown to suppress foam cell formation. TMAO can also contribute to atherosclerosis by inhibiting reverse cholesterol transport [115] and by inducing atherosclerosis-promoting inflammatory proteins in vascular cells[122]. In addition to atherosclerosis, increased levels of TMAO are also associated with a frequency of thrombotic events and platelet activation[123].
Although many studies have reported associations between plasma levels of choline, TMAO, and cardiovascular disease [114, 116, 124, 125] the role of TMAO in disease development is still under debate. A recent study comparing conventional and GF Apoe knockout mice fed diets with or without choline supplementation found no effect of choline enrichment on aortic root atherosclerosis in mice[126]. TMAO production was dependent on the presence of intestinal bacteria but no relationship between TMAO levels and lesions was observed. These results are in contrast to Wang et al who found a significant correlation between TMAO and lesion size[116]. Similarly, studies using diets supplemented with L-carnitine, which result in increased levels of TMAO, have shown opposite effects on aortic lesions in two different laboratories, further emphasizing context dependency on experimental outcome[115, 127].
5 Clinical interventions for treatment of dyslipidemia and NAFLD targeting the gut microbiota
The gut microbiota has been targeted for treatment of diseases related to dyslipidemia. Strategies include supplementing the diet with fibers to enhance the growth or activity of beneficial bacteria (prebiotics), live bacteria (probiotics) or a combination of pre- and probiotics (symbiotics).
A meta-analysis of 11 minor clinical trials using fermented milk and probiotics show beneficial effects on serum lipid profiles[128]. Another meta-analysis focusing on studies using Lactobacillus formulations found an improvement of total serum cholesterol and LDL, but not triglycerides or HDL[129]. One study found improvement of triglycerides but not cholesterol after short-term co-administration of Bifidobacteriae and Lactobacilli strains in healthy subjects, whereas another study observed a decrease in total cholesterol, LDL and triglycerides as well as an increase in HDL using a Bifidobacterium/yeast extract symbiotic[130]. A recent meta-analysis of studies on treatment of NAFLD targeting the gut microbiota showed an overall reduction of AST and ALT levels when pooling data from 25 clinical trials using pre-, pro or symbiotics [131].
Statins lower cholesterol levels by inhibiting HMG-CoA reductase, but may also exert a lipid-lowering effect through interaction with the gut microbiota. Liu et al demonstrated that the cholesterol-lowering effect of rosuvastatin was reflected in microbial alpha diversity measured after eight weeks of treatment [ Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64. https://doi.org/10.1038/nature18846. Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913-6.e7. doi:10.1053/j.gastro.2012.06.031. Koutnikova H, Genser B, Monteiro-Sepulveda M, Faurie J-M, Rizkalla S, Schrezenmeir J et al. Impact of bacterial probiotics on obesity, diabetes and non-alcoholic fatty liver disease related variables: a systematic review and meta-analysis of randomised controlled trials. 2019;9(3):e017995. doi:10.1136/bmjopen-2017-017995 %J BMJ Open. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. https://doi.org/10.1126/science.1241214. Zhou D, Pan Q, Shen F, Cao HX, Ding WJ, Chen YW, et al. Total fecal microbiota transplantation alleviates high-fat diet-induced steatohepatitis in mice via beneficial regulation of gut microbiota. Sci Rep. 2017;7(1):1529. https://doi.org/10.1038/s41598-017-01751-y. Tremaroli V, Karlsson F, Werling M, Stahlman M, Kovatcheva-Datchary P, Olbers T, et al. Roux-en-Y Gastric Bypass and Vertical Banded Gastroplasty Induce Long-Term Changes on the Human Gut Microbiome Contributing to Fat Mass Regulation. Cell Metab. 2015;22(2):228–38. https://doi.org/10.1016/j.cmet.2015.07.009. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. https://doi.org/10.1038/nature05414. Katsiki N, Mikhailidis DP, Mantzoros CS. Non-alcoholic fatty liver disease and dyslipidemia: An update. Metabolism: clinical and experimental. 2016;65(8):1109–23. https://doi.org/10.1016/j.metabol.2016.05.003. Grundy SM. Metabolic syndrome update. Trends Cardiovasc Med. 2016;26(4):364–73. https://doi.org/10.1016/j.tcm.2015.10.004. Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani Patrice D, Bäckhed F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metabolism. 2015;22(4):658–68. https://doi.org/10.1016/j.cmet.2015.07.026. Just S, Mondot S, Ecker J, Wegner K, Rath E, Gau L, et al. The gut microbiota drives the impact of bile acids and fat source in diet on mouse metabolism. Microbiome. 2018;6(1):134. https://doi.org/10.1186/s40168-018-0510-8. Lam YY, Ha CW, Hoffmann JM, Oscarsson J, Dinudom A, Mather TJ, et al. Effects of dietary fat profile on gut permeability and microbiota and their relationships with metabolic changes in mice. Obesity (Silver Spring). 2015;23(7):1429–39. https://doi.org/10.1002/oby.21122. Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansionand colitis in Il10-/- mice. Nature. 2012;487(7405):104-8. https://doi.org/10.1038/nature11225. Jackman JA, Yoon BK, Li D, Cho NJ. Nanotechnology Formulations for Antibacterial Free Fatty Acids and Monoglycerides. Molecules (Basel, Switzerland). 2016;21(3):305. https://doi.org/10.3390/molecules21030305. Shilling M, Matt L, Rubin E, Visitacion MP, Haller NA, Grey SF, et al. Antimicrobial effects of virgin coconut oil and its medium-chain fatty acids on Clostridium difficile. Journal of medicinal food. 2013;16(12):1079–85. https://doi.org/10.1089/jmf.2012.0303. Sheu CW, Freese E. Effects of fatty acids on growth and envelope proteins of Bacillus subtilis. J Bacteriol. 1972;111(2):516–24. Zheng CJ, Yoo JS, Lee TG, Cho HY, Kim YH, Kim WG. Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS letters. 2005;579(23):5157–62. https://doi.org/10.1016/j.febslet.2005.08.028. Chen P, Torralba M, Tan J, Embree M, Zengler K, Starkel P, et al. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology. 2015;148(1):203–14.e16. https://doi.org/10.1053/j.gastro.2014.09.014. Kim YJ. Liu RH. Increase of Conjugated Linoleic Acid Content in Milk by Fermentation with Lactic Acid Bacteria. 2002;67(5):1731–7. https://doi.org/10.1111/j.1365-2621.2002.tb08714.x. Ogawa J, Kishino S, Ando A, Sugimoto S, Mihara K, Shimizu S. Production of conjugated fatty acids by lactic acid bacteria. Journal of bioscience and bioengineering. 2005;100(4):355–64. https://doi.org/10.1263/jbb.100.355. Brown JM, McIntosh MK. Conjugated linoleic acid in humans: regulation of adiposity and insulin sensitivity. J Nutr. 2003;133(10):3041–6. https://doi.org/10.1093/jn/133.10.3041. Wargent E, Sennitt MV, Stocker C, Mayes AE, Brown L, O'Dowd J, et al. Prolonged treatment of genetically obese mice with conjugated linoleic acid improves glucose tolerance and lowers plasma insulin concentration: possible involvement of PPAR activation. Lipids Health Dis. 2005;4:3. https://doi.org/10.1186/1476-511x-4-3. Granlund L, Juvet LK, Pedersen JI, Nebb HI. Trans10, cis12-conjugated linoleic acid prevents triacylglycerol accumulation in adipocytes by acting as a PPARgamma modulator. J Lipid Res. 2003;44(8):1441–52. https://doi.org/10.1194/jlr.M300120-JLR200. Ecker J, Liebisch G, Patsch W, Schmitz G. The conjugated linoleic acid isomer trans-9,trans-11 is a dietary occurring agonist of liver X receptor alpha. Biochem Biophys Res Commun. 2009;388(4):660–6. https://doi.org/10.1016/j.bbrc.2009.08.048. Coakley M, Ross RP, Nordgren M, Fitzgerald G, Devery R, Stanton C. Conjugated linoleic acid biosynthesis by human-derived Bifidobacterium species. Journal of applied microbiology. 2003;94(1):138–45. Lee HY, Park JH, Seok SH, Baek MW, Kim DJ, Lee KE, et al. Human originated bacteria, Lactobacillus rhamnosus PL60, produce conjugated linoleic acid and show anti-obesity effects in diet-induced obese mice. Biochim Biophys Acta. 2006;1761(7):736–44. https://doi.org/10.1016/j.bbalip.2006.05.007. Miyamoto J, Mizukure T, Park SB, Kishino S, Kimura I, Hirano K, et al. A gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, ameliorates intestinal epithelial barrier impairment partially via GPR40-MEK-ERK pathway. J Biol Chem. 2015;290(5):2902–18. https://doi.org/10.1074/jbc.M114.610733. Goto T, Kim YI, Furuzono T, Takahashi N, Yamakuni K, Yang HE, et al. 10-oxo-12(Z)-octadecenoic acid, a linoleic acid metabolite produced by gut lactic acid bacteria, potently activates PPARgamma and stimulates adipogenesis. Biochem Biophys Res Commun. 2015;459(4):597–603. https://doi.org/10.1016/j.bbrc.2015.02.154. Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104(3):979–84. https://doi.org/10.1073/pnas.0605374104. Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, et al. The gut microbiota modulates host energy and lipid metabolism in mice. Journal of Lipid Research. 2010;51(5):1101–12. https://doi.org/10.1194/jlr.M002774. Rabot S, Membrez M, Bruneau A, Gerard P, Harach T, Moser M, et al. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010:4948–59. https://doi.org/10.1096/fj.10-164921. Kindt A, Liebisch G, Clavel T, Haller D, Hormannsperger G, Yoon H, et al. The gut microbiota promotes hepatic fatty acid desaturation and elongation in mice. Nat Commun. 2018;9(1):3760. https://doi.org/10.1038/s41467-018-05767-4. Schedin-Weiss S, Caesar I, Winblad B, Blom H, Tjernberg LO. Super-resolution microscopy reveals gamma-secretase at both sides of the neuronal synapse. Acta Neuropathol Commun. 2016;4:29. https://doi.org/10.1186/s40478-016-0296-5. Yoo SR, Kim YJ, Park DY, Jung UJ, Jeon SM, Ahn YT, et al. Probiotics L. plantarum and L. curvatus in combination alter hepatic lipid metabolism and suppress diet-induced obesity. Obesity (Silver Spring). 2013;21(12):2571–8. https://doi.org/10.1002/oby.20428. An HM, Park SY, Lee DK, Kim JR, Cha MK. Lee SW et al. Antiobesity and lipid-lowering effects of Bifidobacterium spp. in high fat diet-induced obese rats. Lipids Health Dis. 2011;10:116. https://doi.org/10.1186/1476-511x-10-116. Castaner O, Goday A, Park YM, Lee SH, Magkos F, Shiow STE, et al. The Gut Microbiome Profile in Obesity: A Systematic Review. Int J Endocrinol. 2018;2018:4095789. https://doi.org/10.1155/2018/4095789. Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500(7464):585–8. https://doi.org/10.1038/nature12480. Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–6. https://doi.org/10.1038/nature12506. Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA, et al. The Gut Microbiome Contributes to a Substantial Proportion of the Variation in Blood Lipids. Circulation research. 2015;117(9):817–24. https://doi.org/10.1161/circresaha.115.306807. Koren O, Spor A, Felin J, Fak F, Stombaugh J, Tremaroli V, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4592–8. https://doi.org/10.1073/pnas.1011383107. Karlsson FH, Fak F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. https://doi.org/10.1038/ncomms2266. Emoto T, Yamashita T, Sasaki N, Hirota Y, Hayashi T, So A, et al. Analysis of Gut Microbiota in Coronary Artery Disease Patients: a Possible Link between Gut Microbiota and Coronary Artery Disease. J Atheroscler Thromb. 2016;23(8):908–21. https://doi.org/10.5551/jat.32672. Da Silva HE, Teterina A, Comelli EM, Taibi A, Arendt BM, Fischer SE, et al. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci Rep. 2018;8(1):1466. https://doi.org/10.1038/s41598-018-19753-9. Michail S, Lin M, Frey MR, Fanter R, Paliy O, Hilbush B, et al. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS microbiology ecology. 2015;91(2):1–9. https://doi.org/10.1093/femsec/fiu002. Del Chierico F, Nobili V, Vernocchi P, Russo A, Stefanis C, Gnani D, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology (Baltimore, Md). 2017;65(2):451–64. https://doi.org/10.1002/hep.28572. Hoyles L, Fernández-Real J-M, Federici M, Serino M, Abbott J, Charpentier J, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nature Medicine. 2018;24(7):1070–80. https://doi.org/10.1038/s41591-018-0061-3. Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology (Baltimore, Md). 2016;63(3):764–75. https://doi.org/10.1002/hep.28356. Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017;25(5):1054–62.e5. https://doi.org/10.1016/j.cmet.2017.04.001. Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221–7. https://doi.org/10.1136/gut.28.10.1221. Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiological reviews. 1990;70(2):567–90. https://doi.org/10.1152/physrev.1990.70.2.567. den Besten G, Lange K, Havinga R, van Dijk TH, Gerding A, van Eunen K, et al. Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am J Physiol Gastrointest Liver Physiol. 2013;305(12):G900–10. https://doi.org/10.1152/ajpgi.00265.2013. Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61(2):364–71. https://doi.org/10.2337/db11-1019. Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. https://doi.org/10.1038/ncomms2852. Ge H, Li X, Weiszmann J, Wang P, Baribault H, Chen JL, et al. Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids. Endocrinology. 2008;149(9):4519–26. https://doi.org/10.1210/en.2008-0059. McNelis JC, Lee YS, Mayoral R, van der Kant R, Johnson AM, Wollam J, et al. GPR43 Potentiates beta-Cell Function in Obesity. Diabetes. 2015;64(9):3203–17. https://doi.org/10.2337/db14-1938. Chambers ES, Viardot A, Psichas A, Morrison DJ, Murphy KG, Zac-Varghese SE, et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut. 2015;64(11):1744–54. https://doi.org/10.1136/gutjnl-2014-307913. Robertson MD, Bickerton AS, Dennis AL, Vidal H, Frayn KN. Insulin-sensitizing effects of dietary resistant starch and effects on skeletal muscle and adipose tissue metabolism. Am J Clin Nutr. 2005;82(3):559–67. https://doi.org/10.1093/ajcn.82.3.559. Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008;105(43):16767–72. https://doi.org/10.1073/pnas.0808567105. Alex S, Lange K, Amolo T, Grinstead JS, Haakonsson AK, Szalowska E, et al. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 2013;33(7):1303–16. https://doi.org/10.1128/mcb.00858-12. Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58(7):1509–17. https://doi.org/10.2337/db08-1637. den Besten G, Bleeker A, Gerding A, van Eunen K, Havinga R, van Dijk TH, et al. Short-Chain Fatty Acids Protect Against High-Fat Diet-Induced Obesity via a PPARgamma-Dependent Switch From Lipogenesis to Fat Oxidation. Diabetes. 2015;64(7):2398–408. https://doi.org/10.2337/db14-1213. Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165(6):1332–45. https://doi.org/10.1016/j.cell.2016.05.041. Yamashita H, Fujisawa K, Ito E, Idei S, Kawaguchi N, Kimoto M, et al. Improvement of obesity and glucose tolerance by acetate in Type 2 diabetic Otsuka Long-Evans Tokushima Fatty (OLETF) rats. Bioscience, biotechnology, and biochemistry. 2007;71(5):1236–43. De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1-2):84–96. https://doi.org/10.1016/j.cell.2013.12.016. Nicolucci AC, Hume MP, Martinez I, Mayengbam S, Walter J, Reimer RA. Prebiotics Reduce Body Fat and Alter Intestinal Microbiota in Children Who Are Overweight or With Obesity. Gastroenterology. 2017;153(3):711–22. https://doi.org/10.1053/j.gastro.2017.05.055. Freeland KR, Wolever TM. Acute effects of intravenous and rectal acetate on glucagon-like peptide-1, peptide YY, ghrelin, adiponectin and tumour necrosis factor-alpha. Br J Nutr. 2010;103(3):460–6. https://doi.org/10.1017/s0007114509991863. Wahlstrom A, Sayin SI, Marschall HU, Backhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016;24(1):41–50. https://doi.org/10.1016/j.cmet.2016.05.005. Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284(5418):1365–8. https://doi.org/10.1126/science.284.5418.1365. Mueller M, Thorell A, Claudel T, Jha P, Koefeler H, Lackner C, et al. Ursodeoxycholic acid exerts farnesoid X receptor-antagonistic effects on bile acid and lipid metabolism in morbid obesity. J Hepatol. 2015;62(6):1398–404. https://doi.org/10.1016/j.jhep.2014.12.034. Sayin Sama I, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, et al. Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-beta-muricholic Acid, a Naturally Occurring FXR Antagonist. Cell Metabolism. 2013;17(2):225–35. https://doi.org/10.1016/j.cmet.2013.01.003. Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annual review of physiology. 2006;68:159–91. https://doi.org/10.1146/annurev.physiol.68.033104.152158. Parseus A, Sommer N, Sommer F, Caesar R, Molinaro A, Stahlman M, et al. Microbiota-induced obesity requires farnesoid X receptor. Gut. 2017;66(3):429–37. https://doi.org/10.1136/gutjnl-2015-310283. Fang S, Suh JM, Reilly SM, Yu E, Osborn O, Lackey D, et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med. 2015;21(2):159–65. https://doi.org/10.1038/nm.3760. Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest. 2006;116(4):1102–9. https://doi.org/10.1172/jci25604. Jiang C, **e C, Lv Y, Li J, Krausz KW, Shi J, et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun. 2015;6:10166. https://doi.org/10.1038/ncomms10166. Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun. 2013;4:2384. https://doi.org/10.1038/ncomms3384. Jiang C, **e C, Li F, Zhang L, Nichols RG, Krausz KW, et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest. 2015;125(1):386–402. https://doi.org/10.1172/jci76738. Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484–9. https://doi.org/10.1038/nature04330. Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10(3):167–77. https://doi.org/10.1016/j.cmet.2009.08.001. Broeders EP, Nascimento EB, Havekes B, Brans B, Roumans KH, Tailleux A, et al. The Bile Acid Chenodeoxycholic Acid Increases Human Brown Adipose Tissue Activity. Cell Metab. 2015;22(3):418–26. https://doi.org/10.1016/j.cmet.2015.07.002. Mouzaki M, Wang AY, Bandsma R, Comelli EM, Arendt BM, Zhang L, et al. Bile Acids and Dysbiosis in Non-Alcoholic Fatty Liver Disease. PLoS One. 2016;11(5):e0151829. https://doi.org/10.1371/journal.pone.0151829. Alisi A, Ceccarelli S, Panera N, Prono F, Petrini S, De Stefanis C, et al. Association between Serum Atypical Fibroblast Growth Factors 21 and 19 and Pediatric Nonalcoholic Fatty Liver Disease. PLoS One. 2013;8(6):e67160. https://doi.org/10.1371/journal.pone.0067160. Wojcik M, Janus D, Dolezal-Oltarzewska K, Kalicka-Kasperczyk A, Poplawska K, Drozdz D, et al. A decrease in fasting FGF19 levels is associated with the development of non-alcoholic fatty liver disease in obese adolescents. Journal of pediatric endocrinology & metabolism : JPEM. 2012;25(11-12):1089–93. https://doi.org/10.1515/jpem-2012-0253. Lindor KD, Kowdley KV, Heathcote EJ, Harrison ME, Jorgensen R, Angulo P, et al. Ursodeoxycholic acid for treatment of nonalcoholic steatohepatitis: results of a randomized trial. Hepatology (Baltimore, Md). 2004;39(3):770–8. https://doi.org/10.1002/hep.20092. Leuschner UF, Lindenthal B, Herrmann G, Arnold JC, Rossle M, Cordes HJ, et al. High-dose ursodeoxycholic acid therapy for nonalcoholic steatohepatitis: a double-blind, randomized, placebo-controlled trial. Hepatology (Baltimore, Md). 2010;52(2):472–9. https://doi.org/10.1002/hep.23727. Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet (London, England). 2015;385(9972):956–65. https://doi.org/10.1016/s0140-6736(14)61933-4. Harrison SA, Rinella ME, Abdelmalek MF, Trotter JF, Paredes AH, Arnold HL, et al. NGM282 for treatment of non-alcoholic steatohepatitis: a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet (London, England). 2018;391(10126):1174–85. https://doi.org/10.1016/s0140-6736(18)30474-4. Erridge C, Attina T, Spickett CM, Webb DJ. A high-fat meal induces low-grade endotoxemia: evidence of a novel mechanism of postprandial inflammation. Am J Clin Nutr. 2007;86(5):1286–92. https://doi.org/10.1093/ajcn/86.5.1286. Velasquez OR, Henninger K, Fowler M, Tso P, Crissinger KD. Oleic acid-induced mucosal injury in develo** piglet intestine. Am J Physiol. 1993;264(3 Pt 1):G576–82. https://doi.org/10.1152/ajpgi.1993.264.3.G576. Levels JH, Abraham PR, van den Ende A, van Deventer SJ. Distribution and kinetics of lipoprotein-bound endotoxin. Infect Immun. 2001;69(5):2821–8. https://doi.org/10.1128/IAI.69.5.2821-2828.2001. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–72. https://doi.org/10.2337/db06-1491. Pirlich M, Norman K, Lochs H, Bauditz J. Role of intestinal function in cachexia. Curr Opin Clin Nutr Metab Care. 2006;9(5):603–6. https://doi.org/10.1097/01.mco.0000241671.09676.d8. Manco M, Putignani L, Bottazzo GF. Gut microbiota, lipopolysaccharides, and innate immunity in the pathogenesis of obesity and cardiovascular risk. Endocr Rev. 2010;31(6):817–44. https://doi.org/10.1210/er.2009-0030. Read TE, Harris HW, Grunfeld C, Feingold KR, Kane JP, Rapp JH. The protective effect of serum lipoproteins against bacterial lipopolysaccharide. Eur Heart J. 1993;14 Suppl K:125-9. Eichbaum EB, Harris HW, Kane JP, Rapp JH. Chylomicrons can inhibit endotoxin activity in vitro. J Surg Res. 1991;51(5):413–6. Harris HW, Grunfeld C, Feingold KR, Rapp JH. Human very low density lipoproteins and chylomicrons can protect against endotoxin-induced death in mice. J Clin Invest. 1990;86(3):696–702. https://doi.org/10.1172/JCI114765. Pajkrt D, Doran JE, Koster F, Lerch PG, Arnet B, van der Poll T, et al. Antiinflammatory effects of reconstituted high-density lipoprotein during human endotoxemia. J Exp Med. 1996;184(5):1601–8. https://doi.org/10.1084/jem.184.5.1601. Feingold KR, Funk JL, Moser AH, Shigenaga JK, Rapp JH, Grunfeld C. Role for circulating lipoproteins in protection from endotoxin toxicity. Infect Immun. 1995;63(5):2041–6. Lehr HA, Sagban TA, Ihling C, Zahringer U, Hungerer KD, Blumrich M, et al. Immunopathogenesis of atherosclerosis: endotoxin accelerates atherosclerosis in rabbits on hypercholesterolemic diet. Circulation. 2001;104(8):914–20. https://doi.org/10.1161/hc3401.093153. Michelsen KS, Wong MH, Shah PK, Zhang W, Yano J, Doherty TM, et al. Lack of Toll-like receptor 4 or myeloid differentiation factor 88 reduces atherosclerosis and alters plaque phenotype in mice deficient in apolipoprotein E. Proc Natl Acad Sci U S A. 2004;101(29):10679–84. https://doi.org/10.1073/pnas.0403249101. Kiechl S, Egger G, Mayr M, Wiedermann CJ, Bonora E, Oberhollenzer F, et al. Chronic infections and the risk of carotid atherosclerosis: prospective results from a large population study. Circulation. 2001;103(8):1064–70. https://doi.org/10.1161/01.cir.103.8.1064. McIntyre CW, Harrison LE, Eldehni MT, Jefferies HJ, Szeto CC, John SG, et al. Circulating endotoxemia: a novel factor in systemic inflammation and cardiovascular disease in chronic kidney disease. Clinical journal of the American Society of Nephrology : CJASN. 2011;6(1):133–41. https://doi.org/10.2215/cjn.04610510. Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25(2):187–91. https://doi.org/10.1038/76048. Kiechl S, Lorenz E, Reindl M, Wiedermann CJ, Oberhollenzer F, Bonora E, et al. Toll-like receptor 4 polymorphisms and atherogenesis. N Engl J Med. 2002;347(3):185–92. https://doi.org/10.1056/NEJMoa012673. Ameziane N, Beillat T, Verpillat P, Chollet-Martin S, Aumont MC, Seknadji P, et al. Association of the Toll-like receptor 4 gene Asp299Gly polymorphism with acute coronary events. Arterioscler Thromb Vasc Biol. 2003;23(12):e61–4. https://doi.org/10.1161/01.ATV.0000101191.92392.1D. Mao J-W, Tang H-Y, Zhao T, Tan X-Y, Bi J, Wang B-Y, et al. Intestinal mucosal barrier dysfunction participates in the progress of nonalcoholic fatty liver disease. Int J Clin Exp Pathol. 2015;8(4):3648–58. Mencin A, Kluwe J, Schwabe RF. Toll-like receptors as targets in chronic liver diseases. Gut. 2009;58(5):704–20. https://doi.org/10.1136/gut.2008.156307. Federico A, Dallio M, Godos J, Loguercio C, Salomone F. Targeting gut-liver axis for the treatment of nonalcoholic steatohepatitis: translational and clinical evidence. Translational research : the journal of laboratory and clinical medicine. 2016;167(1):116–24. https://doi.org/10.1016/j.trsl.2015.08.002. Everard A, Geurts L, Caesar R, Van Hul M, Matamoros S, Duparc T, et al. Intestinal epithelial MyD88 is a sensor switching host metabolism towards obesity according to nutritional status. Nat Commun. 2014;5. https://doi.org/10.1038/ncomms6648. Everard A, Geurts L, Caesar R, Van Hul M, Matamoros S, Duparc T, et al. Intestinal epithelial MyD88 is a sensor switching host metabolism towards obesity according to nutritional status. Nat Commun. 2014;5:5648. https://doi.org/10.1038/ncomms6648. Friedman SL. Liver fibrosis in 2012: Convergent pathways that cause hepatic fibrosis in NASH. Nat Rev Gastroenterol Hepatol. 2013;10(2):71–2. https://doi.org/10.1038/nrgastro.2012.256. Wree A, McGeough MD, Pena CA, Schlattjan M, Li H, Inzaugarat ME, et al. NLRP3 inflammasome activation is required for fibrosis development in NAFLD. Journal of molecular medicine (Berlin, Germany). 2014;92(10):1069–82. https://doi.org/10.1007/s00109-014-1170-1. Miele L, Valenza V, La Torre G, Montalto M, Cammarota G, Ricci R, et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology (Baltimore, Md). 2009;49(6):1877–87. https://doi.org/10.1002/hep.22848. Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–84. https://doi.org/10.1056/NEJMoa1109400. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–85. https://doi.org/10.1038/nm.3145. Wang Z, Tang WH, Buffa JA, Fu X, Britt EB, Koeth RA, et al. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur Heart J. 2014;35(14):904–10. https://doi.org/10.1093/eurheartj/ehu002. Bennett BJ, Davis RC, Civelek M, Orozco L, Wu J, Qi H, et al. Genetic Architecture of Atherosclerosis in Mice: A Systems Genetics Analysis of Common Inbred Strains. PLoS genetics. 2015;11(12):e1005711. https://doi.org/10.1371/journal.pgen.1005711. Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem. 2015;290(9):5647–60. https://doi.org/10.1074/jbc.M114.618249. Shih DM, Wang Z, Lee R, Meng Y, Che N, Charugundla S, et al. Flavin containing monooxygenase 3 exerts broad effects on glucose and lipid metabolism and atherosclerosis. J Lipid Res. 2015;56(1):22–37. https://doi.org/10.1194/jlr.M051680. Miao J, Ling AV, Manthena PV, Gearing ME, Graham MJ, Crooke RM, et al. Flavin-containing monooxygenase 3 as a potential player in diabetes-associated atherosclerosis. Nat Commun. 2015;6:6498. https://doi.org/10.1038/ncomms7498. Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17(1):49–60. https://doi.org/10.1016/j.cmet.2012.12.011. Seldin MM, Meng Y, Qi H, Zhu W, Wang Z, Hazen SL, et al. Trimethylamine N-Oxide Promotes Vascular Inflammation Through Signaling of Mitogen-Activated Protein Kinase and Nuclear Factor-kappaB. Journal of the American Heart Association. 2016;5(2). https://doi.org/10.1161/jaha.115.002767. Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell. 2016;165(1):111–24. https://doi.org/10.1016/j.cell.2016.02.011. Mente A, Chalcraft K, Ak H, Davis AD, Lonn E, Miller R, et al. The Relationship Between Trimethylamine-N-Oxide and Prevalent Cardiovascular Disease in a Multiethnic Population Living in Canada. The Canadian journal of cardiology. 2015;31(9):1189–94. https://doi.org/10.1016/j.cjca.2015.06.016. Troseid M, Ueland T, Hov JR, Svardal A, Gregersen I, Dahl CP, et al. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J Intern Med. 2015;277(6):717–26. https://doi.org/10.1111/joim.12328. Lindskog Jonsson A, Caesar R, Akrami R, Reinhardt C, Fak Hallenius F, Boren J, et al. Impact of Gut Microbiota and Diet on the Development of Atherosclerosis in Apoe(-/-) Mice. Arterioscler Thromb Vasc Biol. 2018;38(10):2318–26. https://doi.org/10.1161/atvbaha.118.311233. Collins HL, Drazul-Schrader D, Sulpizio AC, Koster PD, Williamson Y, Adelman SJ, et al. L-Carnitine intake and high trimethylamine N-oxide plasma levels correlate with low aortic lesions in ApoE(-/-) transgenic mice expressing CETP. Atherosclerosis. 2016;244:29–37. https://doi.org/10.1016/j.atherosclerosis.2015.10.108. Shimizu M, Hashiguchi M, Shiga T, Tamura HO, Mochizuki M. Meta-Analysis: Effects of Probiotic Supplementation on Lipid Profiles in Normal to Mildly Hypercholesterolemic Individuals. PLoS One. 2015;10(10):e0139795. https://doi.org/10.1371/journal.pone.0139795. Wu Y, Zhang Q, Ren Y, Ruan Z. Effect of probiotic Lactobacillus on lipid profile: A systematic review and meta-analysis of randomized, controlled trials. PLoS One. 2017;12(6):e0178868. https://doi.org/10.1371/journal.pone.0178868. Ruscica M, Pavanello C, Gandini S, Macchi C, Botta M, Dall'Orto D, et al. Nutraceutical approach for the management of cardiovascular risk - a combination containing the probiotic Bifidobacterium longum BB536 and red yeast rice extract: results from a randomized, double-blind, placebo-controlled study. Nutr J. 2019;18(1):13. https://doi.org/10.1186/s12937-019-0438-2. Loman BR, Hernandez-Saavedra D, An R, Rector RS. Prebiotic and probiotic treatment of nonalcoholic fatty liver disease: a systematic review and meta-analysis. Nutr Rev. 2018;76(11):822–39. https://doi.org/10.1093/nutrit/nuy031. Liu Y, Song X, Zhou H, Zhou X, **a Y, Dong X, et al. Gut Microbiome Associates With Lipid-Lowering Effect of Rosuvastatin in Vivo. Front Microbiol. 2018;9:530. https://doi.org/10.3389/fmicb.2018.00530. Nolan JA, Skuse P, Govindarajan K, Patterson E, Konstantinidou N, Casey PG, et al. The influence of rosuvastatin on the gastrointestinal microbiota and host gene expression profiles. Am J Physiol Gastrointest Liver Physiol. 2017;312(5):G488–G97. https://doi.org/10.1152/ajpgi.00149.2016. Caparros-Martin JA, Lareu RR, Ramsay JP, Peplies J, Reen FJ, Headlam HA, et al. Statin therapy causes gut dysbiosis in mice through a PXR-dependent mechanism. Microbiome. 2017;5(1):95. https://doi.org/10.1186/s40168-017-0312-4. Portugal LR, Goncalves JL, Fernandes LR, Silva HP, Arantes RM, Nicoli JR, et al. Effect of Lactobacillus delbrueckii on cholesterol metabolism in germ-free mice and on atherogenesis in apolipoprotein E knock-out mice. Braz J Med Biol Res. 2006;39(5):629–35. https://doi.org/10.1590/s0100-879x2006000500010. Fak F, Backhed F. Lactobacillus reuteri prevents diet-induced obesity, but not atherosclerosis, in a strain dependent fashion in Apoe-/- mice. PLoS One. 2012;7(10):e46837. https://doi.org/10.1371/journal.pone.0046837. Li J, Lin S, Vanhoutte PM, Woo CW, Xu A. Akkermansia Muciniphila Protects Against Atherosclerosis by Preventing Metabolic Endotoxemia-Induced Inflammation in Apoe-/- Mice. Circulation. 2016;133(24):2434–46. https://doi.org/10.1161/CIRCULATIONAHA.115.019645. Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RR, et al. Daily Sampling Reveals Personalized Diet-Microbiome Associations in Humans. Cell Host Microbe. 2019;25(6):789–802.e5. https://doi.org/10.1016/j.chom.2019.05.005. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell. 2015;163(5):1079–94. https://doi.org/10.1016/j.cell.2015.11.001.References
Acknowledgments
We thank R. Perkins and A. Wahlström for reading and commenting on the manuscript. We also thank A. Hallén for her assistance with figures and artwork. Work in the authors’ laboratory is supported by The Swedish Research Council Formas (2017-02001).
Funding
Open access funding provided by University of Gothenburg.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Schoeler, M., Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev Endocr Metab Disord 20, 461–472 (2019). https://doi.org/10.1007/s11154-019-09512-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11154-019-09512-0