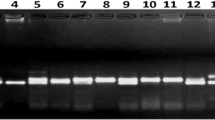
Abstract
Indian mustard is an economically important oilseed crop in India; therefore, exploring the genetic diversity of various germplasm collections is quite relevant for its genetic improvement. In the present study, we investigated genetic diversity and population structure among a panel of 145 Indian mustard germplasm accessions using 11 agro-morphological traits and 182 SSR markers. The result of variance analysis exhibited significant differences among the tested genotypes inferring the presence of a high degree of variability. The phenotypic coefficient of variation for seed yield (CV = 28.26%) was the highest among all the studied traits followed by biological yield (CV = 25.72%). Days to maturity ranged from 130 to 141 days (CV = 1.88%), which was the minimum variation exhibited by Indian mustard genotypes. Out of 235 SSR primer pairs evaluated, 182 (77.45%) SSRs resulted in polymorphic amplicons, while 53 (22.55%) SSRs amplified monomorphic loci. Allele number varied from 2 to 7 with 3.97 average number of alleles per SSR marker. PIC value varied from 0.03 (SJ1668I) to 0.71 (EJU4) with an average value of 0.39 per SSR marker. Gene diversity in the present study ranged from 0.03 (SJ1668I) to 0.75 (EJU4) with an average value of 0.46. Cluster analysis using morphological traits and SSR markers grouped all 145 accessions into two major clusters each, respectively. Population structure analysis grouped all the genotypes into three subpopulations with varying degrees of admixture genotypes. Analysis of molecular variance inferred that 88% of the total variation resides within the groups. The resulting estimates of genetic diversity and population structure may be useful for the effective management of Indian mustard germplasm resources and for future mustard breeding programs.



Similar content being viewed by others
Availability of Data and Materials
All the data and plant material are available with the corresponding author (AKT).
References
Banga S, Kumar PR, Bhajan R, Singh D, Banga SS (2015) Brassica oilseeds: breeding and management. In Kumar A, Banga SS, Meena PD, Kumar PR (eds) CAB International, Oxfordshire, UK, 11–41
Burton WA, Ripley VL, Potts DA, Salisbury PA (2004) Assessment of genetic diversity in selected breeding lines and cultivars of canola quality Brassica juncea and their implications for canola breeding. Euphytica 136:181–192
Chauhan JS, Singh KH, Singh VV, Kumar S (2011) Hundred years of rapeseed-mustard breeding in India: accomplishments and future strategies. Indian J Agri Sci 81(12):1093–1109
Earl DA, vonHoldt BM (2012) Structure harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Ghosh S, Mazumder M, Mondal B, Mukherjee A, De A, Bose R, Das S, Bhattacharyya S, Basu D (2019) Morphological and SSR marker-based genetic diversity analysis of Indian mustard (Brassica juncea L.) differing in Alternaria brassicicola tolerance. Euphytica 215:206. https://doi.org/10.1007/s10681-019-2523-1
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet Res Int. https://doi.org/10.1155/2015/431487
Govt of India (2018) Agricultural statistics at a glance. Directorate of Economics and Statistics, Department of Agriculture and Cooperation, Ministry of Agriculture, Govt. of India, New Delhi. http://cands.daenet.nic.in/PDF. Accessed 21 Jul 2021
Habib MA, Manmohan Sharma P, Salgotra RK, Kiran U (2019) Analysis of allelic differentiation and prediction of suitable parents among Brassica juncea L genotypes using microsatellite markers. Int J Curr Microbiol App Sci 8(6):3071–3081
Huangfu C, Song X, Qiang SG (2009) ISSR variation within and among wild Brassica juncea populations: implication for herbicide resistance evolution. Genet Resour Crop Evol 56:913–924
Ignjatovic-Micic D, Ristic D, Babic V, Andjelkovic V, Vancetovic J (2015) A simple SSR analysis for genetic diversity estimation of maize landraces. Genetika 47:53–62
Jain A, Bhatia S, Banga SS, Prakash S, Lakshmikumaran M (1994) Potential use of random amplified polymorphic DNA (RAPD) technique to study the genetic diversity in Indian mustard (Brassica juncea) and its relationship to heterosis. Theor Appl Genet 88:116–122
Kaur R, Sharma AK, Rani R, Mawlong I, Rai PK (2019) Medicinal qualities of mustard oil and its role in human health against chronic diseases: A review. Asian J Dairy Food Res 382:98–104
Khan MA, Rabbani MA, Muhammad M, Ajmal SK, Malik MA (2008) Assessment of genetic variation within Indian mustard germplasm using RAPD markers. J Interg Plant Biol 50:385–392
Kumar N, Avtar R, Ahlawat N, Rakesh (2020) Molecular diversity analysis using simple sequence repeats (SSRs) in ‘A’ and ‘R’ lines of Ogura CMS system in Indian mustard. Int J Chem Stud 8(4):1276-1282
Kumari A, Kumari A (2018) Studies on genetic diversity in Indian mustard (Brassica Juncea Czern & Coss) for morphological characters under changed climate in the mid-hills of Himalayas. Pharma Innov J 7(7):290–296
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129
Mir JI, Islam I, Kudesia R (2015) Evaluation of genetic diversity in Brassica juncea (L.) using protein profiling and molecular marker (RFLP). Int J Plant Breeding Genet 9(2):77–85
Pandey S, Ansari WA, Pandey M, Singh B (2018) Genetic diversity of cucumber estimated by morpho-physiological and EST-SSR markers. Physiol Mol Biol Plants 24(1):135–146. https://doi.org/10.1007/s12298-017-0489-9
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. http://darwin.cirad.fr/darwin. Accessed 21 Jul 2021
Pritchard JK, Wen W (2003) Documentation for the structure software, version 2.3.4. Department of Human Genetics, University of Chicago, Chicago. http://pritch.bsd.uchicago.edu/software. Accessed 21 Jul 2021
Qi X, Yang J, Zhang M (2008) AFLP-based genetic diversity assessment among Chinese vegetable mustard (Brassica juncea (L.) Czern.). Genet Resour Crop Evol 55:705–711
Rabbani MA, Iwabuchi A, Murakami Y, Suzuki T, Takayanagi K (1998) Genetic diversity in mustard (Brassica juncea L.) germplasm from Pakistan as determined by RAPDs. Euphytica 103(2):235–242
Saini N, Rai AN (2020) Molecular analysis of diversity present in Brassica juncea genotypes with the help of SSR markers. Int J Curr Microbiol App Sci 9(07):2987–2993. https://doi.org/10.20546/ijcmas.2020.907.352
Santos RC, Pires JL, Correa RX (2012) Morphological characterization of leaf, flower, fruit and seed traits among Brazilian Theobroma species. Genet Resour Crop Evol 59:327–345
Sharma D, Nanjundan J, Singh L, Singh SP, Parmar N, SujithKumar MS, Singh KH, Mishra AK, Singh R, Verma KS, Thakur AK (2020) Genetic diversity in leafy mustard (Brassica juncea var. rugosa) as revealed by agro-morphological traits and SSR markers. Physiol Mol Biol Plants. https://doi.org/10.1007/s12298-020-00883-2
Shyam C, Tripathi MK, Tiwari S, Tripathi N, Ahuja A (2020) Molecular characterization and identification of Brassica genotype(s) for low and high erucic acid content using SSR markers. Global J BioSci Biotechnol 9(2):56–66
Singh BK, Thakur AK, Tiwari SK, Siddiqui SA, Singh VV, Rai PK (2012) Transferability of Brassica-derived microsatellites to related genera and their implications for phylogenetic analysis. Natl Acad Sci Lett 35(1):37–44
Singh KH, Misra AK, Kumar A (2006) Draft guidelines to conduct the tests for distinctness, uniformity and stability in rapeseed and mustard (Brassica). National Research Centre on Rapeseed Mustard, Bharatpur, Rajasthan, India
Singh KH, Shakya R, Thakur AK, Chauhan DK, Chauhan JS (2013) Assessment of genetic diversity in Indian mustard (Brassica juncea L Czern & Coss) using morphological traits and RAPD markers. Natl Acad Sci Lett 36(4):419–427
Singh VK, Avtar R, Mahavir NK, Manjeet RK, Rathore V (2020) Assessment of genetic relationship among diverse Indian mustard (Brassica juncea L.) genotypes using XLSTAT. Electronic J Plant Breeding 11(2):674–680. https://doi.org/10.37992/2020.1102.109
Srivastava A, Gupta V, Pental D, Pradhan AK (2001) AFLP based genetic diversity assessment amongst agronomically important natural and some newly synthesized lines of Brassica juncea. Theor Appl Genet 102:193–199
Sudan J, Khajuria P, Gupta SK, Singh R (2016) Analysis of molecular diversity in Indian and exotic genotypes of Brassica juncea using SSR markers. Indian J Genet 76(3):361–364. https://doi.org/10.5958/0975-6906.2016.00054.7
Tahira R, Ihsan-Ullah, Saleem M (2013) Evaluation of genetic diversity of raya (Brassica juncea) through RAPD markers. Int J Agric Biol 15:1163–1168
Tamanna A, Khan AU (2005) Map** and analysis of simple sequence repeats in the Arabidopsis thaliana genome. Bioinformation 1:64–68
Thakur AK, Parmar N, Singh KH, Nanjundan J (2020a) Current achievements and future prospects of genetic engineering in Indian mustard (Brassica juncea L. Czern & Coss.). Planta 252:56. https://doi.org/10.1007/s00425-020-03461-8
Thakur AK, Singh BK, Verma V, Chauhan JS (2013) Direct organogenesis in Brassica juncea var. NRCDR-2 and analysis of genetic uniformity using RAPD markers. Natl Acad Sci Lett 36:403–409
Thakur AK, Singh KH, Lal S, Nanjundan J, Yasin JK, Singh D (2018) SSR marker variations in Brassica species provide insight into the origin and evolution of Brassica amphidiploids. Hereditas 155:6. https://doi.org/10.1186/s41065-017-0041-5
Thakur AK, Singh KH, Parmar N, Sharma D, Mishra DC, Singh L, Nanjundan J, Yadav S (2020b) Population structure and genetic diversity as revealed by SSR markers in Ethiopian mustard (Brassica carinata A. Braun): a potential edible and industrially important oilseed crop. Genet Resour Crop Evol. https://doi.org/10.1007/s10722-020-00988-3
Thakur AK, Singh KH, Singh L, Nanjundan J, Khan YJ, Singh D (2017) Patterns of subspecies genetic diversity among oilseed Brassica rapa as revealed by agro-morphological traits and SSR markers. J Plant Biochem Biotechnol 26(3):282–292
Thakur AK, Singh KH, Singh L, Nanjundan J, Rana MK, Singh D (2015) Transferability of SSR markers of Brassica species to some popular varieties of Brassica juncea. Proc Natl Acad Sci India Sect B Biol Sci 85(4):1001–1010. https://doi.org/10.1007/s40011-014-0486
Vinu V, Singh N, Vasodev S, Yadav DK, Kumar S, Naresh S, Bhat SR, Prabhu KV (2013) Assessment of genetic diversity in Brassica juncea (Brassicaceae) genotypes using phenotypic differences and SSR markers. Rev Biol Trop 61(4):1919–1934
Yadav M, Rana JS (2012) ISSR markers assisted studies on genetic diversity in Brassica juncea. Int J Pharma Bio Sci 3(2):402
Yadava DK, Parida SK, Dwivedi SK, Varshney A, Ghazi IA, Sujata V, Mohapatra T (2009) Cross-transferability and polymorphic potential of genomic STMS markers of Brassica Species. J Plant Biochem Biotechnol 18(1):29–36
Yang et al (2016) The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet 48:1225–1232. https://doi.org/10.1038/ng.3657
Yousuf M, Bhat TM, Kudesia R (2013) Comparative genetic diversity studies in mustard (Brassica juncea) varieties using randomly amplified polymorphic DNA (RAPD) analysis. African J Biotechnol 12(22):3430–3434
Zhu S, Zhang X, Liu Q, Luo T, Tang Z, Zhou Y (2018) The genetic diversity and relationships of cauliflower (Brassica oleracea var. botrytis) inbred lines assessed by using SSR markers. PLoS ONE 13(12):e0208551. https://doi.org/10.1371/journal.pone.0208551
Acknowledgements
The authors are thankful to the Director, ICAR-Directorate of Rapeseed-Mustard Research, Bharatpur, Rajasthan, for providing funding to carry out this research work. The authors are also thankful to Dr. S.S. Banga, National Professor, PAU, Ludhiana, Punjab, for providing B-genome-specific SSR markers.
Author information
Authors and Affiliations
Contributions
AKT, KHS, and JN conceived the idea, developed the work plan, and edited the manuscript. DS, LS, and NP performed the field and lab experiments. KSV helped in the manuscript writing. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
Genetic diversity in a panel of 145 Indian mustard germplasm accessions was analyzed using agro-morphological traits and SSR markers, which has helped in the identification of genetically diverse lines to be used as donors in future breeding programs aimed at Indian mustard genetic improvement.
Supplementary Information
Below is the link to the electronic supplementary material.
11105_2022_1339_MOESM1_ESM.tiff
Supplementary file1 (TIFF 64 kb). Fig. S1 Graph showing the number of subpopulations in Indian mustard germplasm panel based on Structure analysis
Rights and permissions
About this article
Cite this article
Sharma, D., Nanjundan, J., Singh, L. et al. Genetic Diversity and Population Structure Analysis in Indian Mustard Germplasm Using Phenotypic Traits and SSR Markers. Plant Mol Biol Rep 40, 579–594 (2022). https://doi.org/10.1007/s11105-022-01339-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-022-01339-5