Abstract
The secular debate on the origin of life on our planet represents one of the open challenges for the scientific community. In this endeavour, chemistry has a pivotal role in disclosing novel scenarios that allow us to understand how the formation of simple organic molecules would be possible in the early primitive geological ages of Earth. Amino acids play a crucial role in biological processes. They are known to be formed in experiments simulating primitive conditions and were found in meteoric samples retrieved throughout the years. Understanding their formation is a key step for prebiotic chemistry. Following this reasoning, we performed a computational investigation over 100′000 structural isomers of natural amino acids. The results we have found suggest that natural amino acids are among the most thermodynamically stable structures and, therefore, one of the most probable ones to be synthesised among their possible isomers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The reason behind the beginning of life on Earth is a widely debated topic, which encompasses many interdisciplinary fields. Chemistry has always played a vital role in explaining how the emergence of biochemical building blocks such as amino acids, nucleotides and sugars could have been possible.
The pioneering work of Oparin (Oparin 1924) suggested how a long period of abiotic synthesis of organic molecules could support the heterotrophic hypothesis of the origin of life on our planet. This idea culminated in the Urey-Miller experiment, where it was proved how the production of the first simple organic compounds could have been possible in the primitive conditions of Earth in the Archean Age (Miller 1953, 1955). In this well-known experiment, the emergence of biotic molecules has been observed in a simulation of the young Earth, taking into account the presence of spark discharge, inorganic molecules, and reducing atmosphere.
Amino acids, amines, hydroxy acids, aldehydes, nitriles, and many more compounds have been identified in the first promising experimental proofs of Oparin’s “primordial soup”. It has been debated for a long time if the Earth atmosphere was likely to be reducing, as proposed in these pioneering works, or a neutral gas mixture. Later studies have analysed in more detail the geochemical condition of the Earth (Holland 1962; Abelson 1966): it was later understood that the early atmosphere was most likely a neutral gas mixture mainly composed of CO2 (1–100 bar) with ∼1 bar N2 (Kasting and Catling 2003).
Moreover, the low yields of amino acids synthesised by spark discharges have raised the criticism that the formation of amino acids needed additional external support (Johnson et al. 2008).The mechanisms of synthesis of amino acids such as ammonium thiocyanate, thiourea, and thioacetamide are unlikely to be obtained in a non-reducing atmosphere. In fact, the synthesis of such amino acids may follow one of these mechanisms: i) amino nitriles and hydroxy nitriles are produced in aqueous solution by aldehydes and HCN produced by the discharge in the atmosphere (known as Strecker’s mechanism); ii) amino and hydroxy acids are a product of the electric sparks by ions and free radicals (Shneur and Ottesen 1966). Lacking significant percentages of oxygen (Kasting and Catling 2003; Catling and Claire 2005), the direct abiotic synthesis of amino acids may have still occurred during the early rise of atmospheric O2 on our planet. Several mechanisms have been suggested to support this idea: the Bucherer–Bergs reaction (Bucherer and Lieb 1934; Taillades et al. 1998), the hydrolysis of hydrogen cyanide oligomers (Oró and Kamat 1961; Ferris et al. 1978), and the Strecker synthesis (Miller 1955). Even if methane and ammonia have been proved necessary to the formation of amino acids (Schlesinger and Miller 1983; Miyakawa et al. 2002), the production of amino acids and nucleotides highly depends on the hydrogen cyanide that is formed in the atmosphere (Ferris et al. 1978). The experimental absence of polymers derived from hydrogen cyanide and the high level of energies required to form non-α-amino acids suggest that no mechanism can be sufficient by itself.
Among the suggested solutions of this conundrum, hydrothermal synthesis may (Wächtershäuser 1988; Steele et al. 2012) have enhanced prebiotic organic evolution, especially in the production of high-yielding compounds derived from cyanates. The oxidation inhibitors in the oceans may have enhanced the formation of organic compounds in a neutral atmosphere. A more suggestive, but sound hypothesis relies on the contributions of extraterrestrial organic compounds to endogenous synthesis (Chyba and Sagan 1992). It has been experimentally proven that complex organic compounds (e.g. amino acids, purines, pyrimidines) exist in carbonaceous meteorites and are indigenous, as in the Murchison meteorite. The Murchison meteorite is a remarkable case, that greatly advanced the study of geochemical properties of meteorites and comets (Hayes 1967; Nagy 1973). Its fall in September 1969 in Murchison (Australia) proved the existence of extraterrestrially formed organic molecules (Kvenvolden et al. 1970).
Specialists in cosmochemistry and astrobiology have kept a position of consensus in the last years regarding the important hel** role of extraterrestrial molecules on the emergence of life on Earth. According to recent studies (Bernstein et al. 1999), life arose quite rapidly, probably helped by complex organic compounds in fallen comets and meteorites and the existence of a wide variety of organic compounds formed by solar ultraviolet irradiation or tholins supports this theory. Early studies on Murchison meteorite have identified the presence of several aromatic hydrocarbons (Oró et al. 1971; Pering and Ponnamperuma 1971; Studier et al. 1972; Levy et al. 1973). The indigenous origin of these molecules is supported by the similar percentages of enantiomers of type D and of type L in compounds such as valine, proline, and more (Cronin and Pizzarello 1983). Some families of amino acids have further put the research on the trail of prebiotic synthesis of peptides and polynucleotides (Cheng et al. 2004).
On the other hand, one of the cornerstones of prebiotic chemistry relies on the chemical stability of natural α-amino acids. The stability, temperature decomposition and temperature range of synthesis of the natural amino acids was recently studied for glycine, cysteine, aspartic acid, asparagine, glutamic acid, glutamine, arginine, and histidine (Weiss et al. 2018) with mass spectrometry. Different hypotheses were made regarding the reasons behind the selection of the 20 natural structures that emerged among all the possible α-amino acids. Recent studies evaluate the role of the 20 canonical α-amino acids in stabilising the protein structure and stability following different criteria (e.g., type of atoms, solubility, biosynthetic cost) (Doig 2017), or tackling this problem computationally (Bywater 2018). These works draw a series of convincing hypothesis why other classes of α-amino acids were not selected, despite the bountiful number of positive mutations that can be induced incorporating them in a protein structure (Li et al. 2018). Experimental evidence was found for the preference of the D-enantiomer of RNA, which lead to the formation of the L- α-amino acids (Bolik et al. 2007). In general, in the literature, the stability of natural α-amino acids over their structural isomers is widely accepted (Luisi 2006), but the scientific literature paradoxically lacks evidence supporting this hypothesis.
Starting from these considerations, we decided to test the stability of the α-amino acids exploring this scientific challenge from another perspective. We investigated the relative stability of the 20 natural α-amino acids compared to their structural isomers, starting from the hypothesis that among all the isomers possible, only the most stable can be likely related to the early emergence of life in prebiotic conditions. The stability of natural α-amino acids should indeed be confronted using an energetic bias with other isomeric structures sharing the same structural formula, which are expected to be less relevant or marginalised and therefore less produced under prebiotic conditions.
In this work, we analysed ca. 100′000 structural isomers of specific amino acids, studying their geometry and their energetic stability by computational methods. In this work, we aimed at answering two questions. The first one is whether among the possible stable isomers that can be computed starting from an amino acid structure, the ones with an amino acid moiety could rank among the more stable ones. The second is whether the natural amino acid structure could be the more stable one, when the α amino acid (i.e., HOOC-C-N) subunit is fixed as a structural constraint of the isomers that can be generated.
Results and Discussion
Computational Methods
The Structural MOlecular Generation (SMOG) program was used to generate isomers starting from the molecular formulas of natural amino acids (Molchanova et al. 1996). To avoid structures that cannot correspond to actual stable compounds (such as strained structures or molecules containing unstable linkages), we employed a set of constraints. In particular, the following functionalities were avoided: strained cycles (cycles with triple bonds, in some cases three members cyclic compounds or more than seven membered rings); unstable moieties or easily hydrolysable groups (N–N single and double bonds, peroxides, ketenes, geminal amino groups, geminal hydroxyl groups, N-oxides, hemiacetals). To prove our hypothesis, less stringent rules were implemented for a selected number of amino acid isomers (see below), while for the remaining ones, the α-amino acid moiety was imposed to reduce the possible number of structures. All structures were optimised in gas phase. This choice was dictated to avoid bias introduced by the implicit solvent model chosen and by the polarity of the medium. Consequently, all amino acid structures were generated and computed in their neutral form (i.e., protonated acid and neutral amine). In this way we aimed to model an environment more like the prebiotic / extra-terrestrial conditions. On the other hand, the absence of an explicit, nucleophilic solvent in the medium able to hydrolise structures such as acetals was considered when ranking the isomers regarding their energetic stability (vide infra).
Hydrogens were added, and 3D structures were built from the two-dimensional ones produced by SMOG by using HyperChem 8.0.10. The software was also used to perform a coarse optimisation via its molecular mechanics MM + routine. The cartesian coordinates generated by this first optimisation step were then re-optimised at the parametrised semi-empirical PM3 level (Stewart 1989a, b), as implemented in Gaussian 09 Rev.B01. This classic semi-empirical method uses formalism based on the Neglect of Differential Diatomic Overlap, allowing very fast and accurate geometry optimisations for small organic molecules. A subset of the optimised structures (vide infra) was subsequently optimised with Density Functional Theory (DFT) (Hohenberg and Kohn 1964; Kohn and Sham 1965) at the ωB97X-D/TZVP level (Schäfer et al. 1992; Chai and Head-Gordon 2008), as implemented in Gaussian 16, Rev.B01. All stationary points were characterised by computing the respective Hessian matrix, probing the absence of imaginary frequencies. All the calculation sequences were batch-executed, and data analysis pipelines were created to automatize collection, extraction, and processing of results.
Some Considerations
An important point was whether to consider the neutral or ionic form of the amino acid. In fact, amino acids in water exist primarily as ionic (zwitterion) form. In the absence of water or other ions, they cannot exist in monomeric form or at least this is not true for all the amino acids (Price et al. 1997; Wyttenbach et al. 2000).
Moreover, the isoelectric point is different from case to case, and this will raise the question of the choice of the value of pH. By bearing this in mind, and since our work was related to the discussion of the amino acid stability regardless of their generation (thus not excluding a random formation from a high energy mixture of atoms such as that existing in plasma or interstellar medium), we decided not to include the zwitterion form. Additionally, if we consider that only amino acids could exist in the zwitterionic form, this should lead to a higher stabilization of amino acids with respect to their isomers, going in the same direction we observed.
Preliminary Calculations
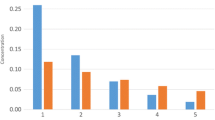
As it is shown in Fig. 1, the relative energy distribution among the isomers generated by our method can change from amino acid to amino acid. This could be related to the presence of different classes of molecules (e.g., acids, amines, carbonates etc.) generated, which directly depends on the formula of the original amino acid and the constraints imposed.
Due to the large number of calculations to perform, only the best candidates were optimised with computational quantum mechanical modelling starting from the PM3 structures. Preliminary quantum calculations based on the Density Functional Theory (DFT) showed that there is a correlation between PM3 and DFT energies (see Fig. 2). For this reason, PM3 energies could be used for the choice of the best candidates; only the lower energies isomers were thus re-optimised by DFT.
Comparison of the electronic energy difference between the optimised geometries of the different isomers of aspartic acid (Asp) relative to their most stable form calculated at the DFT (vertical axis) and PM3 (horizontal axis) levels. The energies are reported in Hartree units. It is evident a direct correlation between the ordering of the isomers obtained from the two different methods
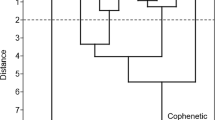
Figure 3 represents a typical output, in this case for threonine (13), where no constraints (vide supra) were imposed. As it is visible from the figure, most of the intermediates will be readily hydrolysed in water. Isomers1, 3 and 7 are hemiacetals, 2, 4, 6, 8, 10 and 12 are α amino alcohols and 5, 9 and 11 are N-substituted carbamic acid derivatives (Dijkstra et al. 2007). Since, on the other hand, it is assumed that liquid water is essential for the emergence of life, only the non-hydrolysable isomers were considered. A recent paper advanced the hypothesis that lipid-encapsulated polymers could be synthesised and directed to form protocells of wet-dry-moist cycles (Damer and Deamer 2020). Peptides are hypothesised to avoid hydrolysis via integration in bilayer structures facilitating the protection against low pH, high temperatures and high cationic concentrations in the hot springs.
It has also to be noted that, in modern metabolism, carbamates are essential intermediates in post-translational modifications (carbamylation) of amino acids (Linthwaite et al. 2018). They are known to spontaneously formed in prebiotic conditions (Preiner et al. 2019; Wimmer et al. 2021), also in the presence of water (do Nascimento Vieira et al. 2020). Consequently, their presence among the most energetically stable isomers formed in the atomic permutation performed is an additional verification of the validity of our computational approach.
As for the optimised structures shown in Fig. 3, after the exclusion of α-amino alcohols, hemiacetals and N-substituted carbamic acids, the real threonine (13) can be considered the first stable molecule. The relative position for the natural α-amino acids after this treatment is reported in Table 1, along with the absolute position obtained by sole energy evaluation.
Relative Energy of Natural Amino Acids
For a selected number of amino acids (viz. Glycine, Threonine, Serine and Alanine), the α-amino acid moiety was not imposed in the structural constraints (vide supra), and all the possible atom permutations leading to stable compounds (e.g., avoid strained structures, vide supra) were calculated. Removing this constraint allows one to determine if the α-amino acid group could be considered relatively more stable to other subunits (e.g., acetals etc.…) and justify constraining its presence in the subsequent calculations. After inspection of these four specific cases, the structures corresponding to the natural amino acids ranked in the first position or a few kcal/mol higher in energy with respect to the most stable isomer, among the possible ones that could be generated (see column Relative Position in Table 1 for Gly, Thr, Ser and Ala). This result might be quite surprising, especially when thousands of possible structures were examined (i.e., for the specific case of Threonine). For this reason, we considered these results plausible proof for the enhanced stability of the α-amino acid moiety. Thus, the α-amino acid group was not permutated for all the other amino acids during the structure generation. This initial benchmark also reduces dramatically the number of isomers to be calculated.
The isomeric leucine and isoleucine represent a particular exception in our study. While isoleucine is the most stable isomer found among the structures generated from its formula, leucine is second in the stability ranking of its isomers despite the limited number of structures generated (see Table 1). Inspecting the most stable structure in Leu, we observed that the most stable structure generated corresponded to isoleucine, which can be generated starting from leucine itself.
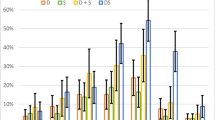
As shown in Table 1, the number of isomers calculated may vary from a few to a hundred thousand. This discrepancy is not only related to the constraints applied to the system and the number of atoms present in the molecule, but also to the index of hydrogen deficiency (IDH) (also called degree of unsaturation) of the amino acid. IDH gives the number of unsaturation present in the structure. All the natural amino acids have an IDH of at least 1, due to the presence of a carbon–oxygen double bond in their carboxylic group. If an additional phenyl ring is present, the IDH value is equal to 5 (i.e., 3 double bonds + 1 ring + C = O). At high IDH values, the number of possible isomers increases dramatically due to all the possible combinations of rings, double and triple bonds. So, in the case of amino acids containing phenyl rings (Phe, Tyr and Trp), we expected many possible structures. We can estimate the number of possible isomers by looking at the elemental analysis. If carbon is more represented, the IDH is greater. Figure 4 shows the position of the natural amino acids in a ternary (CON) diagram.
Natural amino acids seem to cluster according to their elemental composition into two groups. As a hypothesis this could be related to the change of the environment (presence or absence of oxygen), but the discussion is beyond the aim of the present study.
Tryptophan was selected as a case example of a compound with high degree of unsaturation. The number of total isomers for Trp becomes computable by imposing the constraints mentioned in the Computational Methods section above and the α-amino acid group. After excluding easily hydrolysable species, the natural compound is the 30th most stable isomer, almost 5 kcal/mol higher in energy from the most stable. This result is extremely interesting, considering that almost 100′000 possible isomers were calculated. By checking the structures of more stable isomers of Trp, we noticed that some of the compounds possess a sp2 carbon directly linked to α-amino acid residue. we think that the metabolic synthesis of Trp and its relative stability to other compounds might play an important role in prebiotic chemistry. It is interesting to notice that Trp was one of the latest amino acids incorporated into the genetic code. While different contradicting theories were developed to rationalise this peculiar event, its late occurrence has drawn unanimous consensus (Davis 2002; Trifonov 2004; Wong 2005; José et al. 2009, 2011; Palacios-Pérez and José 2019). Albeit a direct correlation between these hypotheses and our energetic results cannot be unequivocally drawn, it is striking that while for the majority of cases the α-amino acid form is the most stable among their isomers, Trp stands out being in the 55th absolute position (although starting from 97,406 structures).
For the isomer calculation of the other two amino acids containing phenyl rings (Phe and Tyr), the presence of the phenyl ring was constrained in the step of isomer generation. This decision was taken both to avoid the formation of a high number of polyenic structures as in Trp, and due to the well-known stability of the phenyl group.
Finally, proline, containing a secondary amine group, was an exception in the way we constrained the α-amino acid subunit. For this molecule, the CON skeleton of α-amino acid was imposed, but without fixing the position and number of hydrogens linked to the C and N. Again, the natural amino acid position is at the top, even if competitors (see Fig. 5) are plausible compounds which do not possess a free COOH group.
Conclusions
The approach presented in this work could give some clues on the stability of natural amino acids. Extensive calculations like those proposed here are accessible nowadays via available supercomputers and batch scripting. As a summary, some interesting findings can be listed even if they are not conclusive proof. First of all, in the examples calculated, the α-amino acid moiety is the most stable. This could be rather surprising, considering that no solvation energy was calculated, a factor which might further increase the amino-acid stabilisation. Furthermore, in most examples, the natural amino acid is the most stable isomer after the removal of hydrolysable compounds. As a matter of fact, the relative energy of the natural amino acid is less than 5–6 kcal/mol higher with respect to the most stable computed isomer. This suggests that, while liquid water or the presence of nucleophiles could play a role in the selection of plausible prebiotic compounds, the natural amino acid structure ranks among the most stable ones generated by our approach.
The stability of natural amino acid has been assessed also for tryptophan, where a large number of isomers was calculated. These findings support the idea that metabolism can increase the overall free energy efficiency and use natural amino acids because they are more stable with respect to other isomers possessing the same structural formula. In a future work, if we presume a totally random combination of atoms, another approach could be related to the selection of elemental composition and number of atoms (molecule complexity). In fact, by checking amino acids found in extraterrestrial bodies or prebiotic experiments, often the most representative amino acids are the simplest. This obviously opens a discussion on which amino acids formed first, depending on their complexity or elemental composition, but this topic will not be discussed here.
In conclusion, computational evaluation of thermodynamical stability can be used in prebiotic chemistry for screening possible intermediates and structural isomers. This approach is ideal for the study of prebiotic chemical reactions networks.
References
Abelson PH (1966) Chemical events on the primitive earth. Proc Natl Acad Sci 55:1365–1372. https://doi.org/10.1073/pnas.55.6.1365
Bernstein MP, Sandford SA, Allamandola LJ (1999) Life’s far-flung raw materials. Sci Am 281:42–49. https://doi.org/10.1038/scientificamerican0799-42
Bolik S, Rübhausen M, Binder S et al (2007) First experimental evidence for the preferential stabilization of the natural D- over the nonnatural L-configuration in nucleic acids. RNA 13:1877–1880. https://doi.org/10.1261/rna.564507
Bucherer HT, Lieb VA (1934) Über die BildungsubstituierterHydantoineausAldehyden und Ketonen. Synthese Von Hydantoinen J fürPraktChemie 141:5–43. https://doi.org/10.1002/prac.19341410102
Bywater RP (2018) Why twenty amino acid residue types suffice(d) to support all living systems. PLoS One 13:e0204883. https://doi.org/10.1371/journal.pone.0204883
Catling DC, Claire MW (2005) How earth’s atmosphere evolved to an oxic state: A status report. Earth Planet Sci Lett 237:1–20. https://doi.org/10.1016/j.epsl.2005.06.013
Chai JD, Head-Gordon M (2008) Long-range corrected hybrid density functionals with damped atom-atom dispersion corrections. Phys Chem Chem Phys 10:6615–6620. https://doi.org/10.1039/b810189b
Cheng CM, Liu XH, Li YM et al (2004) N-phosphoryl amino acids and biomolecular origins. Review paper in honor of the 50th anniversary of the publication of “A production of amino acids under possible primitive earth conditions” (Miller, 1953). Orig Life EvolBiosph 34:455–464. https://doi.org/10.1023/B:ORIG.0000043122.97856.79
Chyba C, Sagan C (1992) Endogenous production, exogenous delivery and impact-shock synthesis of organic molecules: An inventory for the origins of life. Nature 355:125–132
Cronin JR, Pizzarello S (1983) Amino acids in meteorites. Adv Sp Res 3:5–18. https://doi.org/10.1016/0273-1177(83)90036-4
Damer B, Deamer D (2020) The hot spring hypothesis for an origin of life. Astrobiology 20:429–452. https://doi.org/10.1089/ast.2019.2045
Davis BK (2002) Molecular evolution before the origin of species. Prog Biophys Mol Biol 79:77–133
Dijkstra ZJ, Doornbos AR, Weyten H et al (2007) Formation of carbamic acid in organic solvents and in supercritical carbon dioxide. J Supercrit Fluids 41:109–114. https://doi.org/10.1016/j.supflu.2006.08.012
do Nascimento Vieira A, Kleinermanns K, Martin WF, Preiner M (2020) The ambivalent role of water at the origins of life. FEBS Lett 594:2717–2733
Doig AJ (2017) Frozen, but no accident – why the 20 standard amino acids were selected. FEBS J 284:1296–1305. https://doi.org/10.1111/febs.13982
Ferris JP, Joshi PC, Edelson EH, Lawless JG (1978) HCN: A plausible source of purines, pyrimidines and amino acids on the primitive earth. J Mol Evol 11:293–311. https://doi.org/10.1007/BF01733839
Hayes JM (1967) Organic constituents of meteorites-a review. GeochimCosmochim Acta 31:1395–1440. https://doi.org/10.1016/0016-7037(67)90019-1
Hohenberg P, Kohn W (1964) Inhomogeneous electron gas. Phys Rev 136:B864. https://doi.org/10.1103/PhysRev.136.B864
Holland HD (1962) Model for the evolution of the earth’s atmosphere. In: Engle EJ, Jaes HL, Leonard BF (eds) Petrologic studies. Geological Society of America, USA, pp 447–477
Johnson AP, Cleaves HJ, Dworkin JP et al (2008) The Miller volcanic spark discharge experiment. Science 322:404. https://doi.org/10.1126/science.1161527
José MV, Govezensky T, García JA, Bobadilla JR (2009) On the evolution of the standard genetic code: Vestiges of critical scale invariance from the RNA world in current prokaryote genomes. PLoS One 4:4340. https://doi.org/10.1371/journal.pone.0004340
José MV, Morgado ER, Govezensky T (2011) Genetic Hotels for the Standard Genetic Code: Evolutionary Analysis Based upon Novel Three-Dimensional Algebraic Models. Bull Math Biol 73:1443–1476. https://doi.org/10.1007/s11538-010-9571-y
Kasting JF, Catling D (2003) Evolution of a habitable planet. In: Annual review of astronomy and astrophysics. Annual Reviews 4139 El Camino Way, P.O. Box 10139, Palo Alto, CA 94303–0139, USA, pp 429–463
Kohn W, Sham LJ (1965) Self-consistent equations including exchange and correlation effects. Phys Rev 140:A1133. https://doi.org/10.1103/PhysRev.140.A1133
Kvenvolden K, Lawless J, Pering K et al (1970) Evidence for extraterrestrial amino-acids and hydrocarbons in the murchison meteorite. Nature 228:923–926
Levy RL, Grayson MA, Wolf CJ (1973) The organic analysis of the murchison meteorite. GeochimCosmochim Acta 37:467–483. https://doi.org/10.1016/0016-7037(73)90212-3
Li JC, Liu T, Wang Y et al (2018) Enhancing protein stability with genetically encoded noncanonical amino acids. J Am Chem Soc 140:15997–16000. https://doi.org/10.1021/jacs.8b07157
Linthwaite VL, Janus JM, Brown AP et al (2018) The identification of carbon dioxide mediated protein post-translational modifications. Nat Commun 9:1–11. https://doi.org/10.1038/s41467-018-05475-z
Luisi PL (2006) The emergence of life: From chemical origins to synthetic biology. Cambridge University Press
Miller SL (1953) A production of amino acids under possible primitive earth conditions. Science 117:528–529. https://doi.org/10.1126/science.117.3046.528
Miller SL (1955) Production of some organic compounds under possible primitive earth conditions. J Am Chem Soc 77:2351–2361. https://doi.org/10.1021/ja01614a001
Miyakawa S, Yamanashi H, Kobayashi K et al (2002) Prebiotic synthesis from CO atmospheres: Implications for the origins of life. Proc Natl Acad Sci U S A 99:14628–14631. https://doi.org/10.1073/pnas.192568299
Molchanova MS, Shcherbukhin VV, Zefirov NS (1996) Computer generation of molecular structures by the SMOG program. J Chem Inf Comput Sci 36:888–899. https://doi.org/10.1021/ci950393z
Nagy B (1973) Carbonaceous meteorites. In: Kopal Z, Cameron AGW (eds) Developments in solar system and space science 1. Elsevier
Oparin AI (1924) Proiskhozhdenie Zhizni Izd. Moskovskii Rabochii, Moscow, 1924; “The Origin of Life,” 2nd Ed., Dover Publications, New York, 1953.
Oró J, Gibert J, Lichtenstein H et al (1971) Amino-acids, aliphatic and aromatic hydrocarbons in the murchison meteorite. Nature 230:105–106. https://doi.org/10.1038/230105a0
Oró J, Kamat SS (1961) Amino-acid synthesis from hydrogen cyanide under possible primitive earth conditions. Nature 190:442–443. https://doi.org/10.1038/190442a0
Palacios-Pérez M, José MV (2019) The evolution of proteome: from the primeval to the very dawn of LUCA. Biosystems 181:1–10. https://doi.org/10.1016/j.biosystems.2019.04.007
Pering KL, Ponnamperuma C (1971) Aromatic hydrocarbons in the murchison meteorite. Science 173:237–239. https://doi.org/10.1126/science.173.3993.237
Preiner M, Xavier JC, Do Nascimento Vieira A et al (2019) Catalysts, autocatalysis and the origin of metabolism. Interface Focus 9
Price WD, Jockusch RA, Williams ER (1997) Is arginine a zwitterion in the gas phase? J Am Chem Soc 119:11988–11989. https://doi.org/10.1021/ja9711627
Schäfer A, Horn H, Ahlrichs R (1992) Fully optimized contracted Gaussian basis sets for atoms Li to Kr. J Chem Phys 97:2571–2577. https://doi.org/10.1063/1.463096
Schlesinger G, Miller SL (1983) Prebiotic synthesis in atmospheres containing CH4, CO, and CO2 - I. Amino Acids J Mol Evol 19:376–382. https://doi.org/10.1007/BF02101642
Shneur EA, Ottesen EA (1966) Extraterrestrial life: an anthology and bibliography. National Academy of Sciences
Steele A, McCubbin FM, Fries M et al (2012) A reduced organic carbon component in martian basalts. Science 337:212–215. https://doi.org/10.1126/science.1220715
Stewart JJP (1989a) Optimization of parameters for semiempirical methods I. Method J Comput Chem 10:209–220. https://doi.org/10.1002/jcc.540100208
Stewart JJP (1989b) Optimization of parameters for semiempirical methods II. Applications J Comput Chem 10:221–264. https://doi.org/10.1002/jcc.540100209
Studier MH, Hayatsu R, Anders E (1972) Origin of organic matter in early solar system-V. Further studies of meteoritic hydrocarbons and a discussion of their origin. GeochimCosmochim Acta 36:189–215. https://doi.org/10.1016/0016-7037(72)90006-3
Taillades J, Beuzelin I, Garrel L et al (1998) N-carbamoyl-α-amino acids rather than free α-amino acids formation in the primitive hydrosphere: A novel proposal for the emergence of prebiotic peptides. Orig Life EvolBiosph 28:61–77. https://doi.org/10.1023/A:1006566810636
Trifonov EN (2004) The triplet code from first principles. J Biomol Struct Dyn 22:1–11. https://doi.org/10.1080/07391102.2004.10506975
Wächtershäuser G (1988) Before enzymes and templates: theory of surface metabolism. Microbiol Rev 52:452–484. https://doi.org/10.1128/mr.52.4.452-484.1988
Weiss IM, Muth C, Drumm R, Kirchner HOK (2018) Thermal decomposition of the amino acids glycine, cysteine, aspartic acid, asparagine, glutamic acid, glutamine, arginine and histidine. BMC Biophys 11:1–15. https://doi.org/10.1186/s13628-018-0042-4
Wimmer JLE, Vieira ADN, Xavier JC et al (2021) The autotrophic core: an ancient network of 404 reactions converts H2, CO2, and NH3 into amino acids, bases, and cofactors. Microorganisms 9:1–16. https://doi.org/10.3390/microorganisms9020458
Wong JTF (2005) Coevolution theory of genetic code at age thirty. BioEssays 27:416–425. https://doi.org/10.1002/bies.20208
Wyttenbach T, Witt M, Bowers MT (2000) On the stability of amino acid zwitterions in the gas phase: the influence of derivatization, proton affinity, and alkali ion addition. J Am Chem Soc 122:3458–3464. https://doi.org/10.1021/ja992546v
Acknowledgements
CINECA is strongly acknowledged for the use of their supercomputer facility (HPC project IsC74_COSMO). Istituto Nazionale di FisicaNucleare (INFN) is gratefully acknowledged.
Funding
No funding to be mentioned.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Crespi, S., Vadivel, D., Bellisario, A. et al. Computational Study of the Stability of Natural Amino Acid isomers. Orig Life Evol Biosph 51, 287–298 (2021). https://doi.org/10.1007/s11084-021-09615-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-021-09615-2