Abstract
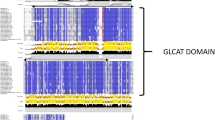
Stevia rebaudiana Bertoni is an important economic crop that is well known for its secondary metabolites, steviol glycosides (SGs), found in leaves. Because the enzymes of deglycosylation (glycoside hydrolases) play important roles in SGs biosynthetic processes, our study is focused on the functions of β-glucosidases in SGs catabolism in stevia. We cloned and characterized 19 stevia GH1 genes based on transcriptomic sequences. The 19 genes were divided into five putative subfamilies in Arabidopsis. Conserved motifs in the SrGH1 proteins were analysed using the online motif-based sequence analysis tool, MEME. Most of the identified proteins contain the conserved ‘TFNEP’ motif (contains the catalytic acid/base) and ‘ITENG’ motif (contains the catalytic nucleophile). Furthermore, the steviol glycoside content and expression of these 19 genes were characterized under constant darkness. The dark treatment lowered the steviol glycoside content significantly, while SrBGLU16 responded to darkness and was markedly upregulated. This study is the first transcriptome-wide analysis of the GH1 family in Stevia rebaudiana. The sequences of 19 SrGH1 members and their expression when grown in darkness were characterized. Among the 19 genes, SrBGLU16 was markedly upregulated by darkness. Thus, we identified SrBGLU16 for further investigation as a possible steviol glycoside beta-glucosidase.



Similar content being viewed by others

References
Goyal SK, Samsher GRK (2010) Stevia (Stevia rebaudiana) a bio-sweetener: a review. Int J Food Sci Nutr 61(1):1–10. https://doi.org/10.3109/09637480903193049
Fengyang L, Yunhe F, Bo L et al (2012) Stevioside suppressed inflammatory cytokine secretion by downregulation of NF-κB and MAPK signaling pathways in LPS-stimulated RAW264.7 cells. Inflammation. 35(5):1669–1675. https://doi.org/10.1007/s10753-012-9483-0
Chatsudthipong V, Muanprasat C (2009) Stevioside and related compounds: therapeutic benefits beyond sweetness. Pharmacol Ther 121(1):41–54. https://doi.org/10.1016/j.pharmthera.2008.09.007
Richman AS, Gijzen M, Starratt AN, Yang Z, Brandle JE (1999) Diterpene synthesis in Stevia rebaudiana: recruitment and up-regulation of key enzymes from the gibberellin biosynthetic pathway. Plant J 19(4):411–421. https://doi.org/10.1046/j.1365-313x.1999.00531.x
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42:D490–D495. https://doi.org/10.1093/nar/gkt1178
Vassão DG, Wielsch N, Gomes AMMM et al (2018) Plant defensive β-glucosidases resist digestion and sustain activity in the gut of a lepidopteran herbivore. Front Plant Sci. 9:1389. https://doi.org/10.3389/fpls.2018.01389
Pankoke H, Buschmann T, Müller C (2013) Role of plant β-glucosidases in the dual defense system of iridoid glycosides and their hydrolyzing enzymes in Plantago lanceolata and Plantago major. Phytochemistry 94:99–107. https://doi.org/10.1016/j.phytochem.2013.04.016
Ketudat Cairns JR, Esen A (2010) β-Glucosidases. Cell Mol Life Sci 67(20):3389–3405. https://doi.org/10.1007/s00018-010-0399-2
Ketudat Cairns JR, Mahong B, Baiya S, Jeon JS (2015) β-Glucosidases: multitasking, moonlighting or simply misunderstood? Plant Sci 241:246–259. https://doi.org/10.1016/j.plantsci.2015.10.014
Yoshiara LY, Madeira TB, de Camargo AC, Shahidi F, Ida EI (2018) Multistep optimization of β-glucosidase extraction from germinated soybeans (Glycine max L. Merril) and recovery of isoflavone aglycones. Foods. 7(7):110. https://doi.org/10.3390/foods7070110
Falcão HG, Handa CL, Silva MBR et al (2018) Soybean ultrasound pre-treatment prior to soaking affects β-glucosidase activity, isoflavone profile and soaking time. Food Chem 269:404–412. https://doi.org/10.1016/j.foodchem.2018.07.028
Zhou Y, Zeng L, Gui J et al (2017) Functional characterizations of β-glucosidases involved in aroma compound formation in tea (Camellia sinensis). Food Res Int 96:206–214. https://doi.org/10.1016/j.foodres.2017.03.049
Wang Z, Wang J, Jiang M et al (2015) Selective production of rubusoside from stevioside by using the sophorose activity of β-glucosidase from Streptomyces sp. GXT6. Appl Microbiol Biotechnol. 99(22):9663–9674. https://doi.org/10.1007/s00253-015-6802-z
Ko JA, Kim YM, Ryu YB et al (2012) Mass production of rubusoside using a novel stevioside-specific β-glucosidase from Aspergillus aculeatus. J Agric Food Chem 60(24):6210–6216. https://doi.org/10.1021/jf300531e
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA (2008) Database indexing for production MegaBLAST searches [published correction appears in Bioinformatics 2008 Dec 15;24(24):2942]. Bioinformatics 24(16):1757–1764. https://doi.org/10.1093/bioinformatics/btn322
Bailey TL, Boden M, Buske FA et al (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Yang YH, Huang SZ, Han YL, Yuan HY, Gu CS, Zhao YH (2014) Base substitution mutations in uridinediphosphate-dependent glycosyltransferase 76G1 gene of Stevia rebaudiana causes the low levels of rebaudioside A: mutations in UGT76G1, a key gene of steviol glycosides synthesis. Plant Physiol Biochem 80:220–225. https://doi.org/10.1016/j.plaphy.2014.04.005
Xu Z, Escamilla-Treviño L, Zeng L et al (2004) Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol Biol 55(3):343–367. https://doi.org/10.1007/s11103-004-0790-1
Opassiri R, Pomthong B, Onkoksoong T, Akiyama T, Esen A, Ketudat Cairns JR (2006) Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 beta-glucosidase. BMC Plant Biol. 6:33. https://doi.org/10.1186/1471-2229-6-33
Gómez-Anduro G, Ceniceros-Ojeda EA, Casados-Vázquez LE et al (2011) Genome-wide analysis of the beta-glucosidase gene family in maize (Zea mays L. var B73). Plant Mol Biol. 77(1–2):159–183. https://doi.org/10.1007/s11103-011-9800-2
Yang Y, Hou M, Zhang T et al (2020) A beta-glucosidase gene from Stevia rebaudiana may be involved in the steviol glycosides catabolic pathway. Mol Biol Rep 47:3577–3584. https://doi.org/10.1007/s11033-020-05450-2
Davies G, Henrissat B (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3:853–859. https://doi.org/10.1016/S0969-2126(01)00220-9
Singh G, Verma AK, Kumar V (2016) Catalytic properties, functional attributes and industrial applications of β-glucosidases. Biotech. https://doi.org/10.1007/s13205-015-0328-z
Fukao T, Yeung E, Bailey-Serres J (2012) The submergence tolerance gene SUB1A delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiol 160(4):1795–1807. https://doi.org/10.1104/pp.112.207738
Yang Y, Huang S, Han Y, Yuan H, Gu C, Wang Z (2015) Environmental cues induce changes of steviol glycosides contents and transcription of corresponding biosynthetic genes in Stevia rebaudiana. Plant Physiol Biochem 86:174–180. https://doi.org/10.1016/j.plaphy.2014.12.004
Yoneda Y, Nakashima H, Miyasaka J, Ohdoi K, Shimizu H (2017) Impact of blue, red, and far-red light treatments on gene expression and steviol glycoside accumulation in Stevia rebaudiana. J Phytochemistry 137:57–65. https://doi.org/10.1016/j.phytochem.2017.02.002
Funding
This study was funded by the Natural Science Foundation of Jiangsu Province (BK20160600), the Young Science Foundation Project of the National Natural Science Foundation of China (31601371 and 31901597).
Author information
Authors and Affiliations
Contributions
Yongheng Yang designed the experiments. Yongheng Yang and Menglan Hou carried out the sequence analysis and gene clone. Menglan Hou carried out the HPLC analysis. **aoyang Xu carried out the RT-PCR. Yongxia Zhang took part in sequence analysis. Yongheng Yang drafted the manuscript. Ting Zhang and Yuming Sun participated and advised in manuscript development. Haiyan Yuan, Haiying Tong and Suzhen Huang advised in manuscript correction. All authors read and approved the final submission.
Corresponding authors
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yang, Y., Zhang, T., Xu, X. et al. Identification of GH1 gene family fgt members in Stevia rebaudiana and their expression when grown in darkness. Mol Biol Rep 47, 8739–8746 (2020). https://doi.org/10.1007/s11033-020-05920-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05920-7