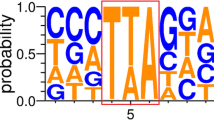
Abstract
Bamboo is one of the most important non-timber forest species in the world, but their molecular breeding lags far behind in contrast to other economic plants. Regarding the difficulties of hybridization and gene modification, the transposon-based insertional mutagenesis might be an alternative, feasible way for molecular breeding of bamboo. A systematic search for potential active transposons identified two full-length mariner-like elements (MLEs) (Ppmar1 and Ppmar2) from moso bamboo in the previous study. Both MLEs contain perfect terminal inverted repeats (TIRs) and a full-length intact transposase. Two transposases contain intact DNA-binding motifs and a DD39D catalytic domain which indicates that Ppmar1 and Ppmar2 are likely active. Here, we deployed a heterologous transposition system of Arabidopsis thaliana to study the transposition activity of Ppmar1 and Ppmar2. The results show that both MLEs could transpose in A. thaliana. Excisions of Ppmar1 and Ppmar2 are usually unperfect as they leave 1–4 bp in excision sites. The reinsertions of both Ppmar1 and Ppmar2 occur at TA dinucleotides and prefer to insert into the TA-rich regions. The insertion sites are dispersed and non-linked. Two active bamboo transposons identified here not only could be applied to construction of the bamboo mutant libraries but also would provide another choice for other plant transposon-based gene tagging.





Similar content being viewed by others
References
Bechtold N, Pelletier G (1998) In planta Agrobacteriummediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82:259–266
Boulin T, Bessereau JL (2007) Mos1-mediated insertional mutagenesis in Caenorhadbitis elegans. Nat Protoc 2:1276–1287
Capy P, Bazin C, Higuet D, Langin T (1998) Dynamics and evolution of transposable elements. Springer-Verlag, Texas
Crénès G, Moundras C, Demattei MV, Bigot Y, Petit A, Renault S (2010) Target site selection by the mariner-like element, Mos1. Genetica 138:509–517
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Dawson A, Finnegan DJ (2003) Excision of the Drosophila mariner transposon Mos1 comparison with bacterial transposition and V (D) J recombination. Mol Cell 11:225–235
Doak TG, Doerder FP, Jahn CL, Herrick G (1994) A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common "D35E" motif. Proc Natl Acad Sci U S A 91:942–946
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Feschotte C, Wessler SR (2002) Mariner-like transposases are widespread and diverse in flowering plants. Proc Natl Acad Sci U S A 99:280–285
Feschotte C, Swamy L, Wessler SR (2003) Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs). Genetics 163:747–758
Fu J (2001) Chinese moso bamboo: its importance. Bamboo 22:5–7
Hartl DL (2001) Discovery of the transposable element mariner. Genetics 157:471–476
Hartl DL, Lohe AR, Lozovskaya ER (1997) Modern thoughts on an ancient mariner: function, evolution, regulation. Annu Rev Genet 31:337–358
Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci U S A 94:2122–2127
Hirochika H (2001) Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol 4:118–122
Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M (1996) Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci U S A 93:7783–7788
Janzen DH (1976) Why bamboos wait so long to flower. Ann Rev Eco Syst 7:347–391
Kolesnik T, Szeverenyi I, Bachmann D, Kumar CS, Jiang S, Ramamoorthy R, Cai M, Ma ZG, Sundaresan V, Ramachandran S (2004) Establishing an efficient Ac/Ds tagging system in rice: large-scale analysis of Ds flanking sequences. Plant J 37:301–314
Kumar CS, Wing RA, Sundaresan V (2005) Efficient insertional mutagenesis in rice using the maize En/Spm elements. Plant J 44:879–892
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Mátés L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, Ma L, Samara-Kuko E, Gysemans C, Pryputniewicz D, Miskey C, Fletcher B, VandenDriessche T, Ivics Z, Izsvák Z (2009) Molecular evolution of a novel hyperactive Slee** Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 41:753–761
Miskey C, Izsvak Z, Plasterk RH, Ivics Z (2003) The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res 31:6873–6881
Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15:1771–1780
Piffanelli P, Droc G, Mieulet D, Lanau N, Bès M, Bourgeois E, Rouvière C, Gavory F, Cruaud C, Ghesquière A, Guiderdoni E (2007) Large-scale characterization of Tos17 insertion sites in a rice T-DNA mutant library. Plant Mol Biol 65(5):587–601
Plasterk RHA, Izsvák Z, Ivics Z (1999) Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet 15:326–332
Scurlocka JMO, Daytonb DC, Hamesb B (2000) Bamboo: an overlooked biomass resource? Biomass Bioenergy 19:229–244
Watanabe M, Ueda K, Manabe I, Akai T (1982) Flowering, seeding, germination, and flowering periodicity of Phyllostachys pubescens. J Jpn For Soc 64:107–111
Yang G, Nagel DH, Feschotte C, Hancock CN, Wessler SR (2009) Tuned for transposition: molecular determinants underlying the hyperactivity of a stowaway MITE. Science 325:1391–1394
Yang G, Weil CF, Wessler SR (2006) A rice Tc1/mariner-like element transposes in yeast. Plant Cell 18:2469–2478
Yant SR, Kay MA (2003) Nonhomologous-end-joining factors regulate DNA repair fidelity during Slee** Beauty element transposition in mammalian cells. Mol Cell Biol 23:8505–8518
Zhou MB, Lu JJ, Zhong H, Tang KX, Tang DQ (2010) Distribution and polymorphism of mariner-like elements in the Bambusoideae subfamily. Plant Syst Evol 289:1–11
Zhou MB, Zhong H, Hu JL, Tang DQ (2015) Ppmar1 and Ppmar2: the first two complete and intact full-length mariner-like elements isolated in Phyllostachys edulis. Acta Botanica Gallica: Botany Letters 162:127–137
Zhou MB, Zhong H, Tang DQ (2011) Isolation and characterization of seventy-nine full-length mariner-like transposases in the Bambusoideae subfamily. J Plant Res 124:607–617
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This work was funded by the grant from the National Natural Science Foundation of China (grant No 31270645 and 31470615) and through the Talents Program of Natural Science Foundation of Zhejiang Province (grant No. LR12C16001).
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary file S1.
The sequences of Ppmar1NA and Ppmar2NA. (FASTA 2 kb)
Supplementary table S2.
The number of resistant transgenic Arabidopsis plants which were selected to investigate the transposition activity of Ppmar1 and Ppmar2. (DOCX 14 kb)
Supplementary figure S3.
The blanking sequences of 10 Ppmar1 insertion sites. The lowercase ta were marked where Ppmar1 inserted, and transposons were represented by the symbols (◄►). (GIF 32 kb)
Supplementary figure S4.
The blanking sequences of 10 Ppmar2 insertion sites. The lowercase ta were marked where Ppmar2 inserted, and transposons were represented by the symbols (◄►). (GIF 33 kb)
Rights and permissions
About this article
Cite this article
Zhou, M., Hu, H., Liu, Z. et al. Two active bamboo mariner-like transposable elements (Ppmar1 and Ppmar2) identified as the transposon-based genetic tools for mutagenesis. Mol Breeding 36, 163 (2016). https://doi.org/10.1007/s11032-016-0588-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-016-0588-2