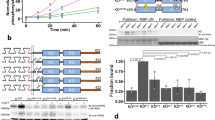
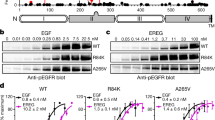
Abstract
Epidermal growth factor receptor (EGFR) dysregulation is observed in many human cancers and is both a cause of oncogenesis and a target for chemotherapy. We previously showed that partial charge neutralization of the juxtamembrane (JX) region of EGFR via the EGFR R1–6 mutant construct induces constitutive receptor activation and transformation of NIH 3T3 cells, both from the plasma membrane and from the ER when combined with the ER-retaining L417H mutation (Bryant et al. in J Biol Chem 288:34930–34942, 2013). Here, we use chemical crosslinking and immunoblotting to show that these mutant constructs form constitutive, phosphorylated dimers in both the plasma membrane and the ER. Furthermore, we combine this electrostatic perturbation with conformationally-restricted receptor mutants to provide evidence that activation of EGFR R1–6 dimers requires functional coupling both between the EGFR extracellular dimerization arms and between intracellular tyrosine kinase domains. These findings provide evidence that the electrostatic charge of the JX region normally serves as a negative regulator of functional dimerization of EGFR.






Similar content being viewed by others
Data Availability
The data, expression plasmids, and cell lines used in this study are available upon reasonable request.
Code Availability
Software application or custom code: not applicable.
Abbreviations
- EGFR:
-
Epidermal growth factor receptor
- ECD:
-
Extracellular domain
- JX:
-
Juxtamembrane
- TKD:
-
Tyrosine kinase domain
- RTK:
-
Receptor tyrosine kinase
- EGFR R1–6:
-
EGFR JX charge-reduced mutant
- EGFR L417H:
-
ER-retained EGFR mutant
- EGFR 246–253*:
-
EGFR ECD domain III mutant that cannot form ECD dimers
- EGFR V948R:
-
EGFR TKD mutant that cannot form asymmetric TKD dimers
- DSS:
-
Disuccinimidyl suberate intracellular crosslinker
- BS3 :
-
Bis(sulfosuccinimidyl)suberate extracellular crosslinker
References
Lemmon MA, Schlessinger J (2010) Cell signaling by receptor tyrosine kinases. Cell 141:1117–1134. https://doi.org/10.1016/j.cell.2010.06.011
Endres NF, Barros T, Cantor AJ, Kuriyan J (2014) Emerging concepts in the regulation of the EGF receptor and other receptor tyrosine kinases. Trends Biochem Sci 39:437–446. https://doi.org/10.1016/j.tibs.2014.08.001
Lemmon M, Schlessinger J, Ferguson KM (2014) The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb Perspect Biol 6:a020768. https://doi.org/10.1101/cshperspect.a020768
Zhang H, Berezov A, Wang Q et al (2007) ErbB receptors: from oncogenes to targeted cancer therapies. J Clin Invest 117:2051–2058. https://doi.org/10.1172/JCI32278
Shan Y, Eastwood MP, Zhang X et al (2012) Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell 149:860–870. https://doi.org/10.1016/j.cell.2012.02.063
Chu CT, Everiss KD, Wikstrand CJ et al (1997) Receptor dimerization is not a factor in the signalling activity of a transforming variant epidermal growth factor receptor (EGFRvIII). Biochem J 324:855–861. https://doi.org/10.1042/bj3240855
Arkhipov A, Shan Y, Das R et al (2013) Architecture and membrane interactions of the EGF receptor. Cell 152:557–569. https://doi.org/10.1016/j.cell.2012.12.030
Sliwkowski MX, Schaefer G, Akita RW et al (1994) Coexpression of erbB2 and er6B3 proteins reconstitutes a high affinity receptor for heregulin. J Biol Chem 269:14661–14665. https://doi.org/10.1002/art.30521
Zhang X, Gureasko J, Shen K et al (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125:1137–1149. https://doi.org/10.1016/j.cell.2006.05.013
Huse M, Kuriyan J (2002) The conformational plasticity of protein kinases. Cell 109:275–282. https://doi.org/10.1016/S0092-8674(02)00741-9
Jura N, Zhang X, Endres NF et al (2011) Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell 42:9–22. https://doi.org/10.1016/j.molcel.2011.03.004
Hubbard SR, Till JH (2000) Protein tyrosine kinase structure and function. Annu Rev Biochem 69:373–398. https://doi.org/10.1146/annurev.biochem.69.1.373
Kancha RK, von Bubnoff N, Duyster J (2013) Asymmetric kinase dimer formation is crucial for the activation of oncogenic EGFRvIII but not for ERBB3 phosphorylation. Cell Commun Signal 11:39. https://doi.org/10.1186/1478-811X-11-39
Hedger G, Sansom MSPP, Koldsø H (2015) The juxtamembrane regions of human receptor tyrosine kinases exhibit conserved interaction sites with anionic lipids. Sci Rep 5:1–10. https://doi.org/10.1038/srep09198
Doerner A, Scheck R, Schepartz A (2015) Growth factor identity is encoded by discrete coiled-coil rotamers in the EGFR juxtamembrane region. Chem Biol 22:776–784. https://doi.org/10.1016/j.chembiol.2015.05.008
He L, Hristova K (2012) Consequences of replacing EGFR juxtamembrane domain with an unstructured sequence. Sci Rep 2:854. https://doi.org/10.1038/srep00854
Pahuja KB, Nguyen TT, Jaiswal BS et al (2018) Actionable activating oncogenic ERBB2/HER2 transmembrane and juxtamembrane domain mutations. Cancer Cell 34:792-806.e5. https://doi.org/10.1016/j.ccell.2018.09.010
Huang Y, Bharill S, Karandur D et al (2016) Molecular basis for multimerization in the activation of the epidermal growth factor receptor. Elife 5:1–27. https://doi.org/10.7554/eLife.14107
Trenker R, Jura N (2020) Receptor tyrosine kinase activation: from the ligand perspective. Curr Opin Cell Biol 63:174–185. https://doi.org/10.1016/j.ceb.2020.01.016
Cai G, Zhu L, Chen X et al (2018) TRAF4 binds to the juxtamembrane region of EGFR directly and promotes kinase activation. Proc Natl Acad Sci USA 115:11531–11536. https://doi.org/10.1073/pnas.1809599115
Xue F, An C, Chen L et al (2019) 4.1B suppresses cancer cell proliferation by binding to EGFR P13 region of intracellular juxtamembrane segment. Cell Commun Signal 17:1–16. https://doi.org/10.1186/s12964-019-0431-6
Mitchell RA, Luwor RB, Burgess AW (2018) Epidermal growth factor receptor: Structure-function informing the design of anticancer therapeutics. Exp Cell Res 371:1–19. https://doi.org/10.1016/j.yexcr.2018.08.009
Gerhart J, Thévenin AF, Bloch E et al (2018) Inhibiting epidermal growth factor receptor dimerization and signaling through targeted delivery of a juxtamembrane domain peptide mimic. ACS Chem Biol 13:2623–2632. https://doi.org/10.1021/acschembio.8b00555
Abd Halim KB, Koldsø H, Sansom MSPP (2015) Interactions of the EGFR juxtamembrane domain with PIP2-containing lipid bilayers: insights from multiscale molecular dynamics simulations. Biochim Biophys Acta - Gen Subj 1850:1017–1025. https://doi.org/10.1016/j.bbagen.2014.09.006
Gambhir A, Hangyás-Mihályné G, Zaitseva I et al (2004) Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys J 86:2188–2207. https://doi.org/10.1016/S0006-3495(04)74278-2
Maeda R, Sato T, Okamoto K et al (2018) Lipid-protein interplay in dimerization of juxtamembrane domains of epidermal growth factor receptor. Biophys J 114:893–903. https://doi.org/10.1016/j.bpj.2017.12.029
Endres NF, Das R, Smith AW et al (2013) Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 152:543–556. https://doi.org/10.1016/j.cell.2012.12.032
Jura N, Endres NF, Engel K et al (2009) Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell 137:1293–1307. https://doi.org/10.1016/j.cell.2009.04.025
Bryant KL, Antonyak MA, Cerione RA et al (2013) Mutations in the polybasic juxtamembrane sequence of both plasma membrane- and endoplasmic reticulum-localized epidermal growth factor receptors confer ligand-independent cell transformation. J Biol Chem 288:34930–34942. https://doi.org/10.1074/jbc.M113.513333
Ward CW, Garrett TPJ (2001) The relationship between the L1 and L2 domains of the insulin and epidermal growth factor receptors and leucine-rich repeat modules. BMC Bioinform 2:4. https://doi.org/10.1186/1471-2105-2-4
Dawson JP, Berger MB, Lin C et al (2005) Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol Cell Biol 25:7734–7742. https://doi.org/10.1128/MCB.25.17.7734-7742.2005
Gosse JJ, Wagenknecht-Wiesner A, Holowka David et al (2005) Transmembrane sequences are determinants of immunoreceptor signaling. J Immunol 175:2123–31
Wakefield DL, Holowka D, Baird B (2017) The FcεRI signaling cascade and integrin trafficking converge at patterned ligand surfaces. Mol Biol Cell 28:3383–3396. https://doi.org/10.1091/mbc.E17-03-0208
Pines G, Köstler WJ, Yarden Y (2010) Oncogenic mutant forms of EGFR: lessons in signal transduction and targets for cancer therapy. FEBS Lett 584:2699–2706. https://doi.org/10.1016/j.febslet.2010.04.019
Chakraborty S, Li L, Puliyappadamba VT et al (2014) Constitutive and ligand-induced EGFR signalling triggers distinct and mutually exclusive downstream signalling networks. Nat Commun 5:5811. https://doi.org/10.1038/ncomms6811
Wang Z, Longo PA, Tarrant MK et al (2011) Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol 18:1388–1393. https://doi.org/10.1038/nsmb.2168
Poger D, Mark AE (2014) Activation of the epidermal growth factor receptor: a series of twists and turns. Biochemistry 53:2710–2721. https://doi.org/10.1021/bi401632z
Valley CC, Arndt-Jovin DJ, Jovin TM et al (2015) Inside-out signaling of oncogenic EGFR mutants promotes ligand-independent dimerization. Biophys J 108:351a. https://doi.org/10.1016/j.bpj.2014.11.1921
Valley CC, Arndt-Jovin DJ, Karedla N et al (2015) Enhanced dimerization drives ligand-independent activity of mutant EGFR in lung cancer. Mol Biol Cell. https://doi.org/10.1091/mbc.E15-05-0269
Cochet C, Kashles O, Chambaz EM et al (1988) Demonstration of epidermal growth factor-induced receptor dimerization in living cells using a chemical covalent cross-linking agent. J Biol Chem 263:3290–3295
Coban O, Zanetti-Dominguez LC, Matthews DR et al (2015) Effect of phosphorylation on EGFR dimer stability probed by single-molecule dynamics and FRET/FLIM. Biophys J 108:1013–1026. https://doi.org/10.1016/j.bpj.2015.01.005
Di Fiore PP, Pierce JH, Fleming TP et al (1987) Overexpression of the human EGF receptor confers an EGF-dependent transformed phenotype to NIH 3T3 cells. Cell 51:1063–1070. https://doi.org/10.1016/0092-8674(87)90592-7
Holowka D, Baird B (2017) Mechanisms of epidermal growth factor receptor signaling as characterized by patterned ligand activation and mutational analysis. Biochim Biophys Acta - Biomembr 1859:1430–1435. https://doi.org/10.1016/j.bbamem.2016.12.015
Acknowledgements
We thank Dr. Maurine Linder (Cornell University) for the use of her VersaDoc MP 5000 imaging system. We appreciate support of these studies by the National Institutes of Health (grants cited elsewhere); the content of this paper is solely the responsibility of the authors.
Funding
This study was supported by the National Institutes of Health (Grant Nos. R01 GM117552 and R01 AI022449).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
NIH 3T3 cells transiently expressing wt-EGFR are constitutively active. NIH 3T3 cells were transiently transfected with wt-EGFR or EGFR R1–6, EGF stimulated (or not) and chemically crosslinked with BS3, lysed, and immunoblotted for EGFR pY1173, total EGFR, and β-actin. Constitutive activation in the absence of EGF (lane 1) is seen in unstimulated cells expressing wt EGFR, likely due to overexpression. The appearance of double band for β-actin is probably due to nonspecific binding to another cell component of slightly higher molecular weight. Supplementary file 1 (TIF 12872 KB)
Online Resource 2
RBL-2H3 cells transiently expressing wt EGFR at low levels do not exhibit constitutive activation. RBL 2H3 cells were transiently transfected with wt-EGFR or EGFR R1–6, EGF stimulated (or not) and chemically crosslinked or not with BS3, lysed, and immunoblotted for EGFR pY1173, total EGFR, and β-actin. There is no constitutive activation in unstimulated cells expressing wt EGFR in the absence of EGF (lane 6); after addition of EGF phosphorylated dimers are trapped by chemical crosslinking (lanes 3-5). Supplementary file 2 (TIF 18105 KB)
Rights and permissions
About this article
Cite this article
Mohr, J.D., Wagenknecht-Wiesner, A., Holowka, D.A. et al. Basic Amino Acids Within the Juxtamembrane Domain of the Epidermal Growth Factor Receptor Regulate Receptor Dimerization and Auto-phosphorylation. Protein J 39, 476–486 (2020). https://doi.org/10.1007/s10930-020-09943-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-020-09943-8